| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
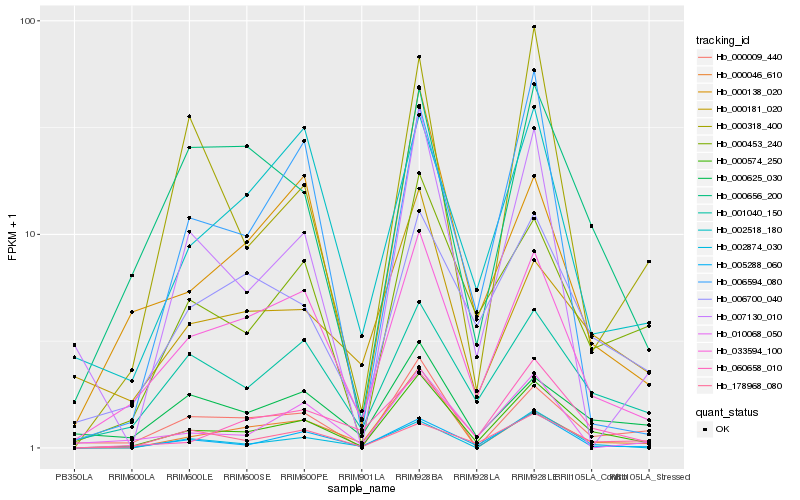
Hb_000574_250 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 2 |
Hb_033594_100 |
0.154641194 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_005288_060 |
0.1557412944 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_000453_240 |
0.1693223165 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 5 |
Hb_060658_010 |
0.1812217448 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 6 |
Hb_001040_150 |
0.1813891964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006700_040 |
0.1843011755 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 8 |
Hb_002874_030 |
0.1931755965 |
- |
- |
hypothetical protein JCGZ_07001 [Jatropha curcas] |
| 9 |
Hb_000009_440 |
0.1975160592 |
- |
- |
PREDICTED: uncharacterized protein LOC105639617 [Jatropha curcas] |
| 10 |
Hb_000656_200 |
0.2024433811 |
- |
- |
PREDICTED: uncharacterized protein LOC105631373 [Jatropha curcas] |
| 11 |
Hb_178968_080 |
0.2025992908 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000138_020 |
0.2110748806 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 13 |
Hb_002518_180 |
0.2147323469 |
- |
- |
hypothetical protein POPTR_0001s25460g [Populus trichocarpa] |
| 14 |
Hb_000318_400 |
0.2150146386 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000625_030 |
0.2156724537 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 16 |
Hb_006594_080 |
0.216896718 |
- |
- |
beta-glucosidase, putative [Ricinus communis] |
| 17 |
Hb_010068_050 |
0.216947691 |
- |
- |
PREDICTED: uncharacterized protein LOC105632963 [Jatropha curcas] |
| 18 |
Hb_000046_610 |
0.2187759379 |
- |
- |
PREDICTED: putative cullin-like protein 2 isoform X3 [Populus euphratica] |
| 19 |
Hb_007130_010 |
0.2198110949 |
- |
- |
- |
| 20 |
Hb_000181_020 |
0.2198456066 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |