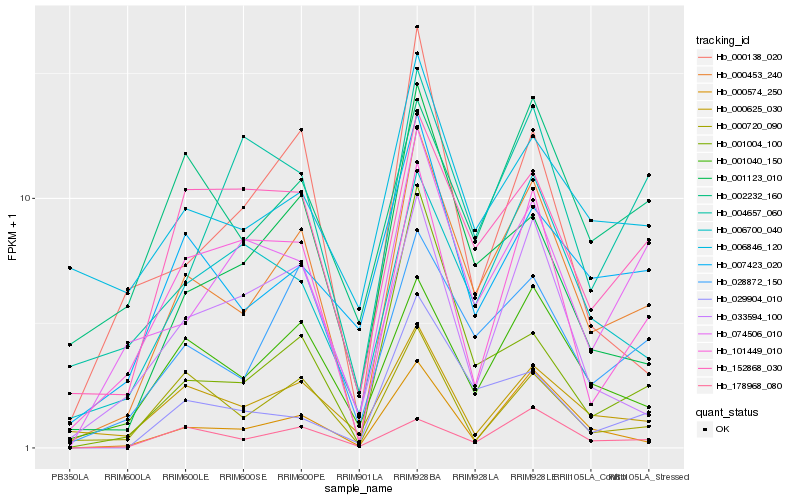
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000453_240 |
0.0 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 2 |
Hb_028872_150 |
0.1333777142 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 3 |
Hb_001040_150 |
0.1533638472 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000574_250 |
0.1693223165 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 5 |
Hb_001123_010 |
0.1702064257 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_152868_030 |
0.1773956592 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 7 |
Hb_006700_040 |
0.1788334957 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 8 |
Hb_007423_020 |
0.1835841017 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 9 |
Hb_033594_100 |
0.1861720992 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable amino-acid acetyltransferase NAGS1, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_029904_010 |
0.1862769574 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 11 |
Hb_074506_010 |
0.1862845017 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 12 |
Hb_178968_080 |
0.1889175138 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000625_030 |
0.1896754996 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 14 |
Hb_101449_010 |
0.1944228214 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 15 |
Hb_000720_090 |
0.1946200154 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 16 |
Hb_001004_100 |
0.1946271958 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_002232_160 |
0.1968722037 |
- |
- |
Transaminase mtnE, putative [Ricinus communis] |
| 18 |
Hb_000138_020 |
0.2001374772 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 19 |
Hb_006846_120 |
0.2029846183 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 20 |
Hb_004657_060 |
0.2056814844 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |