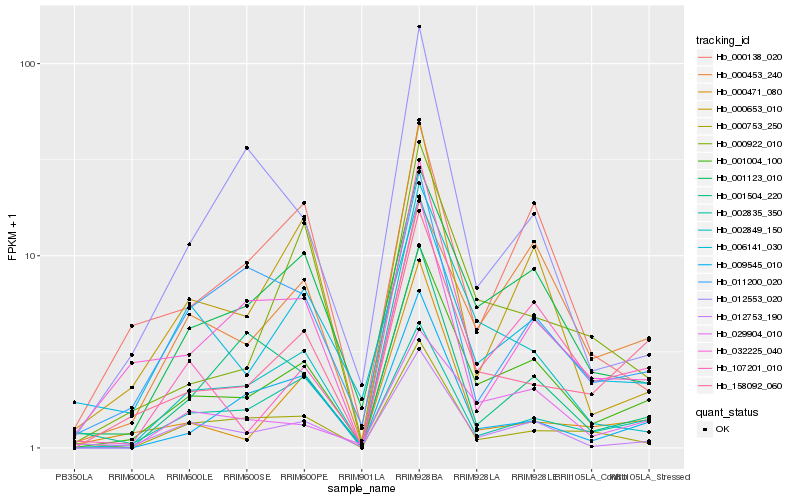
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001004_100 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_001123_010 |
0.1392735524 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_012753_190 |
0.1404322627 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 4 |
Hb_158092_060 |
0.1508179241 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_029904_010 |
0.1655301211 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 6 |
Hb_002849_150 |
0.1725091079 |
- |
- |
hypothetical protein JCGZ_14367 [Jatropha curcas] |
| 7 |
Hb_000653_010 |
0.1734500788 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 8 |
Hb_000922_010 |
0.1829835696 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 9 |
Hb_006141_030 |
0.1876424856 |
- |
- |
- |
| 10 |
Hb_009545_010 |
0.1883511351 |
transcription factor |
TF Family: GNAT |
Acetyltransferase [Medicago truncatula] |
| 11 |
Hb_107201_010 |
0.1889907704 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 12 |
Hb_000453_240 |
0.1946271958 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 13 |
Hb_000138_020 |
0.2019773794 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 14 |
Hb_001504_220 |
0.2071446575 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_011200_020 |
0.208559084 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_000753_250 |
0.2085592181 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 17 |
Hb_002835_350 |
0.2114382157 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 18 |
Hb_032225_040 |
0.2133057873 |
- |
- |
- |
| 19 |
Hb_000471_080 |
0.2148250888 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 20 |
Hb_012553_020 |
0.2177427155 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |