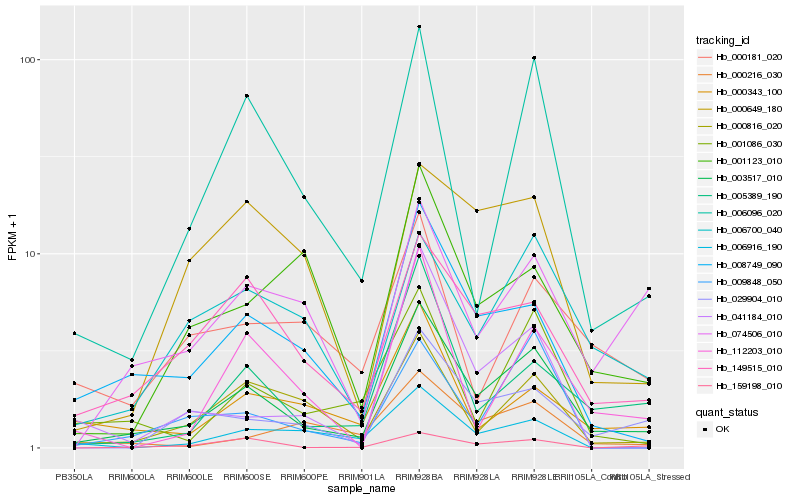
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003517_010 |
0.0 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_008749_090 |
0.1759125592 |
- |
- |
hypothetical protein CICLE_v10017161mg [Citrus clementina] |
| 3 |
Hb_149515_010 |
0.1865964081 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 4 |
Hb_005389_190 |
0.1926878053 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 5 |
Hb_000816_020 |
0.2014453994 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_006096_020 |
0.202584904 |
- |
- |
Os06g0343500 [Oryza sativa Japonica Group] |
| 7 |
Hb_029904_010 |
0.2031078161 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 8 |
Hb_159198_010 |
0.2071227667 |
- |
- |
hypothetical protein VITISV_033829 [Vitis vinifera] |
| 9 |
Hb_001086_030 |
0.2129968141 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 10 |
Hb_006700_040 |
0.2134190591 |
- |
- |
PREDICTED: vacuolar-sorting receptor 6-like [Jatropha curcas] |
| 11 |
Hb_000343_100 |
0.2206878409 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 12 |
Hb_000216_030 |
0.2231272583 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 13 |
Hb_112203_010 |
0.2289019733 |
- |
- |
- |
| 14 |
Hb_074506_010 |
0.229677997 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 15 |
Hb_001123_010 |
0.2362430167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_009848_050 |
0.2403603482 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK13 [Elaeis guineensis] |
| 17 |
Hb_000181_020 |
0.2415677223 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_041184_010 |
0.2424348584 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 19 |
Hb_000649_180 |
0.2436151189 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 20 |
Hb_006916_190 |
0.2438736808 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |