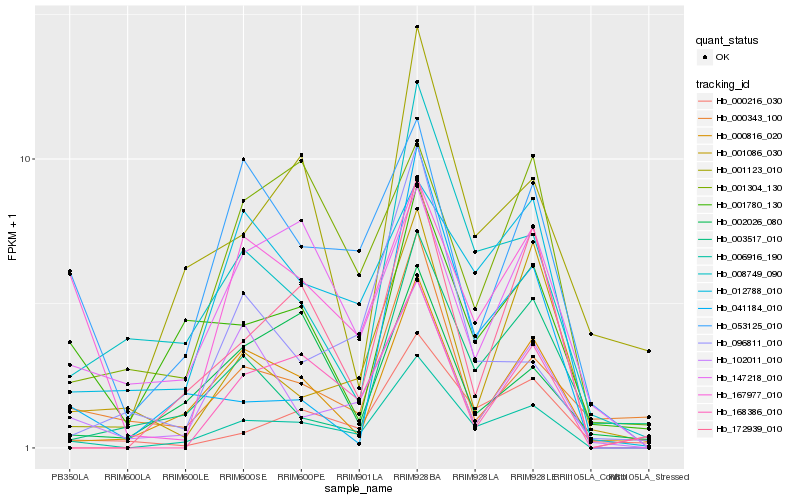
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_190 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_172939_010 |
0.2005024367 |
- |
- |
unknown [Picea sitchensis] |
| 3 |
Hb_000216_030 |
0.2022396234 |
- |
- |
aldehyde oxidase, putative [Ricinus communis] |
| 4 |
Hb_008749_090 |
0.2045521779 |
- |
- |
hypothetical protein CICLE_v10017161mg [Citrus clementina] |
| 5 |
Hb_000816_020 |
0.2095310851 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_012788_010 |
0.2189019253 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 7 |
Hb_168386_010 |
0.221013933 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 8 |
Hb_102011_010 |
0.2236665793 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 9 |
Hb_167977_010 |
0.2290065069 |
- |
- |
hypothetical protein POPTR_0185s002102g, partial [Populus trichocarpa] |
| 10 |
Hb_001780_130 |
0.2301261102 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 11 |
Hb_001123_010 |
0.2328266164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_053125_010 |
0.2345023996 |
- |
- |
PREDICTED: uncharacterized protein LOC104881508 [Vitis vinifera] |
| 13 |
Hb_096811_010 |
0.2386834228 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At1g61190 [Vitis vinifera] |
| 14 |
Hb_003517_010 |
0.2438736808 |
- |
- |
PREDICTED: ABC transporter C family member 3-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000343_100 |
0.2449842295 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 16 |
Hb_147218_010 |
0.2451908633 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 17 |
Hb_002026_080 |
0.2454061451 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 18 |
Hb_001086_030 |
0.2454672228 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 19 |
Hb_041184_010 |
0.2483420861 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 20 |
Hb_001304_130 |
0.2511550744 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like transcription factor PAT1 [Jatropha curcas] |