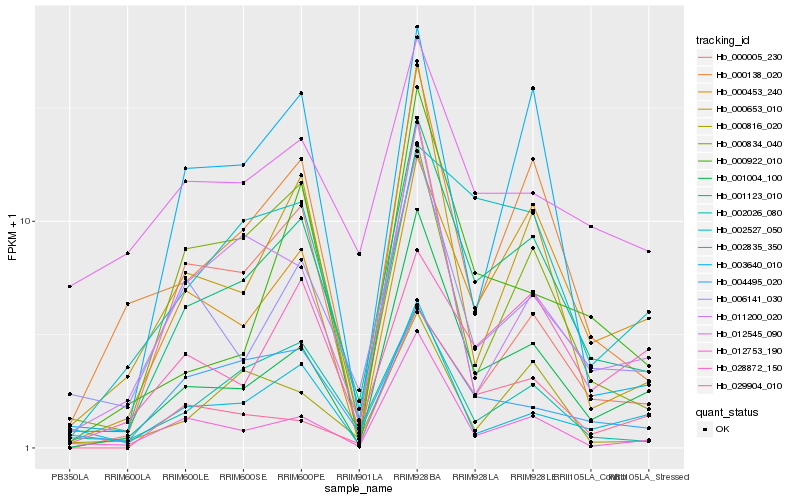
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001123_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001004_100 |
0.1392735524 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000138_020 |
0.1533258885 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 4 |
Hb_006141_030 |
0.164546395 |
- |
- |
- |
| 5 |
Hb_000453_240 |
0.1702064257 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 6 |
Hb_002026_080 |
0.1718595046 |
desease resistance |
Gene Name: TIR_2 |
nucleoside-triphosphatase, putative [Ricinus communis] |
| 7 |
Hb_012753_190 |
0.1779647069 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 8 |
Hb_000653_010 |
0.1824144769 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 9 |
Hb_000816_020 |
0.1943633003 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_029904_010 |
0.1961895547 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 11 |
Hb_002835_350 |
0.1969740511 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 12 |
Hb_000922_010 |
0.1977005802 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 13 |
Hb_000834_040 |
0.1993721854 |
- |
- |
PREDICTED: ras-related protein Rab7 [Cucumis melo] |
| 14 |
Hb_002527_050 |
0.1999801087 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 15 |
Hb_011200_020 |
0.2002984285 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 16 |
Hb_004495_020 |
0.2020214604 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_003640_010 |
0.203465743 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 18 |
Hb_012545_090 |
0.2035521691 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 19 |
Hb_028872_150 |
0.2038835816 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 20 |
Hb_000005_230 |
0.2060621104 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |