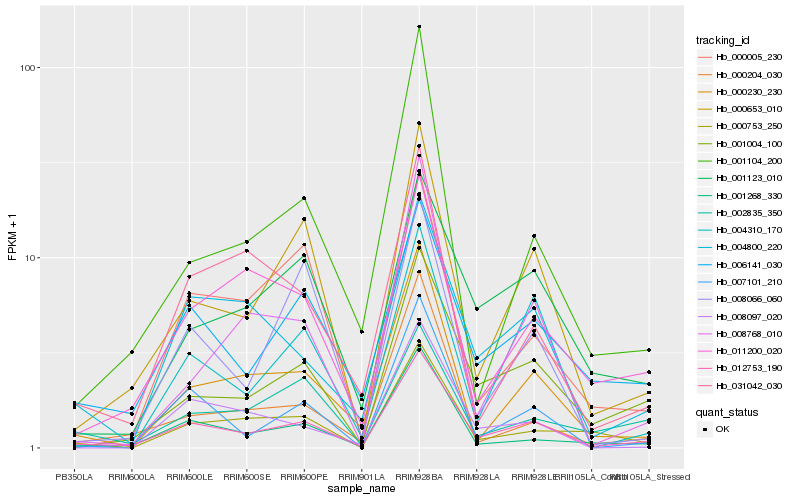
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012753_190 |
0.0 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 2 |
Hb_000653_010 |
0.1347390358 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 3 |
Hb_001004_100 |
0.1404322627 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_006141_030 |
0.1511097063 |
- |
- |
- |
| 5 |
Hb_007101_210 |
0.1512602717 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000230_230 |
0.1547793229 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 7 |
Hb_001268_330 |
0.1567868394 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_004800_220 |
0.1628792146 |
- |
- |
PREDICTED: random slug protein 5 [Jatropha curcas] |
| 9 |
Hb_001123_010 |
0.1779647069 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004310_170 |
0.1805758095 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 11 |
Hb_011200_020 |
0.1814635015 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 12 |
Hb_000753_250 |
0.184102992 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 13 |
Hb_000204_030 |
0.1872598965 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 14 |
Hb_001104_200 |
0.1903610744 |
- |
- |
Phytosulfokine 3 precursor, putative [Theobroma cacao] |
| 15 |
Hb_008097_020 |
0.1911968559 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 16 |
Hb_008066_060 |
0.1914118635 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: subtilisin-like protease SBT1.6 [Jatropha curcas] |
| 17 |
Hb_000005_230 |
0.1925969325 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 18 |
Hb_002835_350 |
0.1928255121 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 19 |
Hb_008768_010 |
0.1939212556 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 20 |
Hb_031042_030 |
0.1949293103 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |