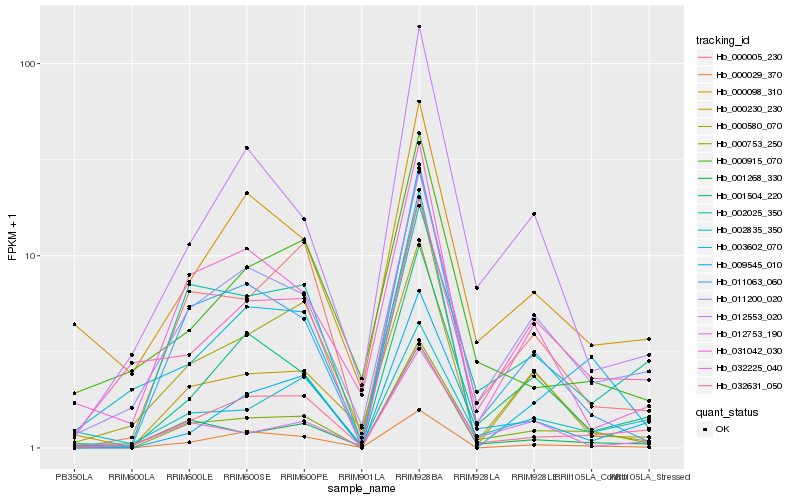
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 2 |
Hb_001268_330 |
0.1559635584 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 3 |
Hb_011200_020 |
0.1601340194 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 4 |
Hb_011063_060 |
0.1665741223 |
- |
- |
PREDICTED: endochitinase-like [Jatropha curcas] |
| 5 |
Hb_000098_310 |
0.16671993 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 6 |
Hb_001504_220 |
0.1682746154 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 7 |
Hb_000230_230 |
0.174988102 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 8 |
Hb_000005_230 |
0.1820393384 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 9 |
Hb_002835_350 |
0.1821407093 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 10 |
Hb_012753_190 |
0.184102992 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 11 |
Hb_003602_070 |
0.191998444 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH93-like [Jatropha curcas] |
| 12 |
Hb_032631_050 |
0.192497065 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_031042_030 |
0.1929048097 |
- |
- |
PREDICTED: WAT1-related protein At5g47470-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_012553_020 |
0.1948176055 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 15 |
Hb_009545_010 |
0.1950930671 |
transcription factor |
TF Family: GNAT |
Acetyltransferase [Medicago truncatula] |
| 16 |
Hb_000915_070 |
0.2001196857 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_032225_040 |
0.2005302783 |
- |
- |
- |
| 18 |
Hb_002025_350 |
0.202469929 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 19 |
Hb_000580_070 |
0.2031858783 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At5g10290 [Jatropha curcas] |
| 20 |
Hb_000029_370 |
0.2045543081 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |