| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
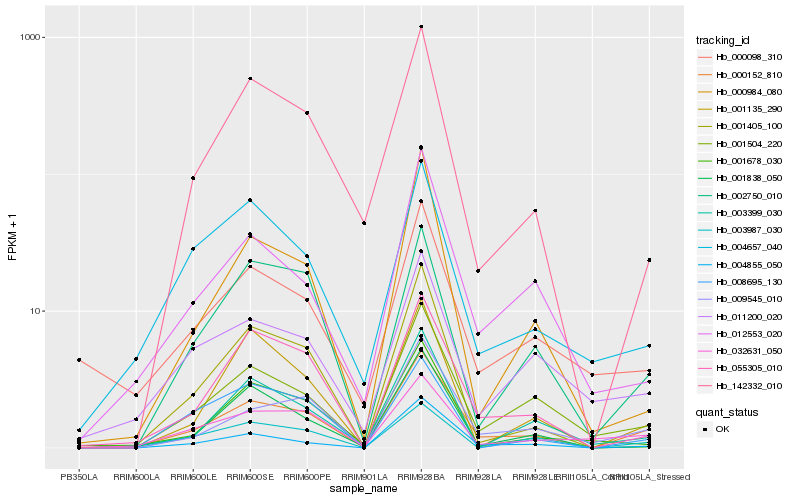
Hb_000152_810 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 2 |
Hb_009545_010 |
0.1388021713 |
transcription factor |
TF Family: GNAT |
Acetyltransferase [Medicago truncatula] |
| 3 |
Hb_001504_220 |
0.1421237018 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 4 |
Hb_055305_010 |
0.1534979453 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 5 |
Hb_004855_050 |
0.153633894 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 6 |
Hb_142332_010 |
0.1714354359 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 7 |
Hb_032631_050 |
0.1734953528 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 8 |
Hb_012553_020 |
0.1750620012 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 9 |
Hb_002750_010 |
0.1803502814 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 10 |
Hb_003399_030 |
0.1834485654 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 11 |
Hb_000984_080 |
0.1846023766 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 12 |
Hb_011200_020 |
0.1851524292 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 13 |
Hb_003987_030 |
0.1870927859 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 14 |
Hb_001838_050 |
0.1890248199 |
- |
- |
hypothetical protein B456_007G218500 [Gossypium raimondii] |
| 15 |
Hb_001135_290 |
0.1908679162 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 16 |
Hb_001405_100 |
0.1914812876 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 17 |
Hb_000098_310 |
0.193203621 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 18 |
Hb_004657_040 |
0.1949511025 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 19 |
Hb_001678_030 |
0.2019405895 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 20 |
Hb_008695_130 |
0.2046377265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |