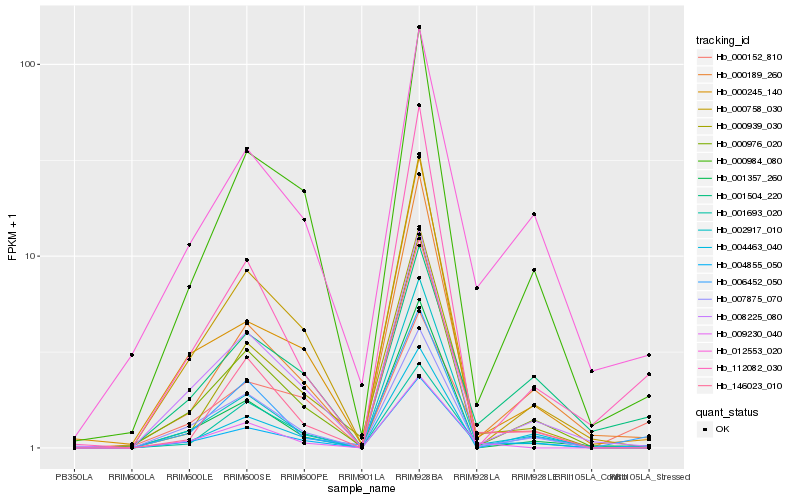
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004855_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 2 |
Hb_012553_020 |
0.1276737683 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 3 |
Hb_007875_070 |
0.127855803 |
- |
- |
hypothetical protein JCGZ_05017 [Jatropha curcas] |
| 4 |
Hb_000984_080 |
0.1383551723 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 5 |
Hb_000976_020 |
0.1385098914 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_008225_080 |
0.1437341694 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 7 |
Hb_000189_260 |
0.1479455183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001504_220 |
0.1522135255 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 9 |
Hb_000152_810 |
0.153633894 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 10 |
Hb_000939_030 |
0.1544772366 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 11 |
Hb_000245_140 |
0.1550485814 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 12 |
Hb_002917_010 |
0.1601807998 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 13 |
Hb_146023_010 |
0.1615726487 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_004463_040 |
0.1617079486 |
- |
- |
- |
| 15 |
Hb_006452_050 |
0.1631854253 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 16 |
Hb_009230_040 |
0.1638802862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000758_030 |
0.1658674646 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 18 |
Hb_001357_260 |
0.1679609398 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_112082_030 |
0.1680898156 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 20 |
Hb_001693_020 |
0.1686964177 |
- |
- |
zinc finger protein, putative [Ricinus communis] |