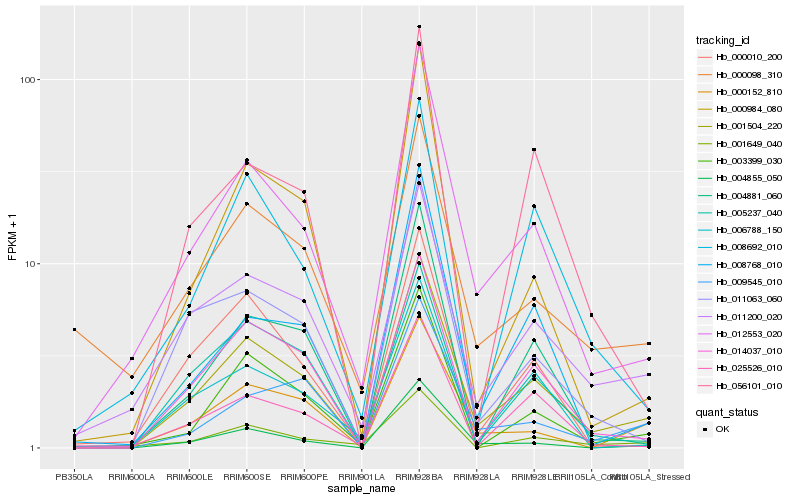
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001504_220 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_012553_020 |
0.107617783 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 3 |
Hb_014037_010 |
0.1204563999 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 4 |
Hb_011200_020 |
0.1261511971 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 5 |
Hb_003399_030 |
0.1388130442 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 6 |
Hb_000152_810 |
0.1421237018 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 7 |
Hb_011063_060 |
0.1492383506 |
- |
- |
PREDICTED: endochitinase-like [Jatropha curcas] |
| 8 |
Hb_004855_050 |
0.1522135255 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 9 |
Hb_004881_060 |
0.1541229562 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 10 |
Hb_009545_010 |
0.1555177847 |
transcription factor |
TF Family: GNAT |
Acetyltransferase [Medicago truncatula] |
| 11 |
Hb_000010_200 |
0.1581848446 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_008692_010 |
0.1582824516 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 13 |
Hb_005237_040 |
0.1587188628 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 14 |
Hb_000098_310 |
0.1589753501 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 15 |
Hb_025526_010 |
0.1598065132 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104452086 [Eucalyptus grandis] |
| 16 |
Hb_001649_040 |
0.1609275424 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 17 |
Hb_008768_010 |
0.1617262768 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 18 |
Hb_056101_010 |
0.16198462 |
- |
- |
hypothetical protein POPTR_0391s00200g [Populus trichocarpa] |
| 19 |
Hb_000984_080 |
0.1625618275 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 20 |
Hb_006788_150 |
0.1631893678 |
- |
- |
conserved hypothetical protein [Ricinus communis] |