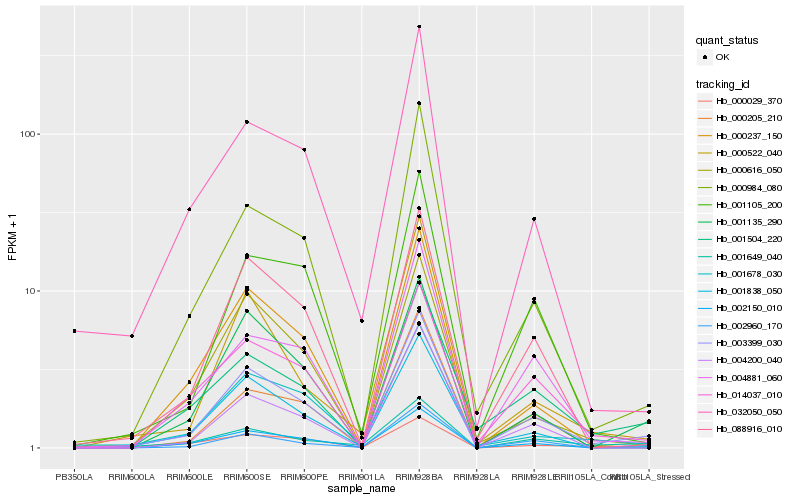
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003399_030 |
0.0 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 2 |
Hb_001838_050 |
0.1175797491 |
- |
- |
hypothetical protein B456_007G218500 [Gossypium raimondii] |
| 3 |
Hb_000984_080 |
0.1238895545 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 4 |
Hb_088916_010 |
0.1266380249 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase At4g25390 [Jatropha curcas] |
| 5 |
Hb_001135_290 |
0.1278279794 |
- |
- |
hypothetical protein JCGZ_06658 [Jatropha curcas] |
| 6 |
Hb_002960_170 |
0.1311028621 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 7 |
Hb_001678_030 |
0.1326179889 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 8 |
Hb_004881_060 |
0.1341508236 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 9 |
Hb_001105_200 |
0.1347730588 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_001504_220 |
0.1388130442 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 11 |
Hb_002150_010 |
0.140833755 |
- |
- |
hypothetical protein POPTR_0001s10240g [Populus trichocarpa] |
| 12 |
Hb_032050_050 |
0.1429676288 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 13 |
Hb_000237_150 |
0.1429844964 |
- |
- |
Epoxide hydrolase 2 [Morus notabilis] |
| 14 |
Hb_004200_040 |
0.1445410776 |
- |
- |
laccase, putative [Ricinus communis] |
| 15 |
Hb_001649_040 |
0.1453978036 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 16 |
Hb_014037_010 |
0.1480628831 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 17 |
Hb_000029_370 |
0.1488034321 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 18 |
Hb_000522_040 |
0.1500351563 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 19 |
Hb_000205_210 |
0.1512081747 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like isoform X1 [Citrus sinensis] |
| 20 |
Hb_000616_050 |
0.1550294059 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |