| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
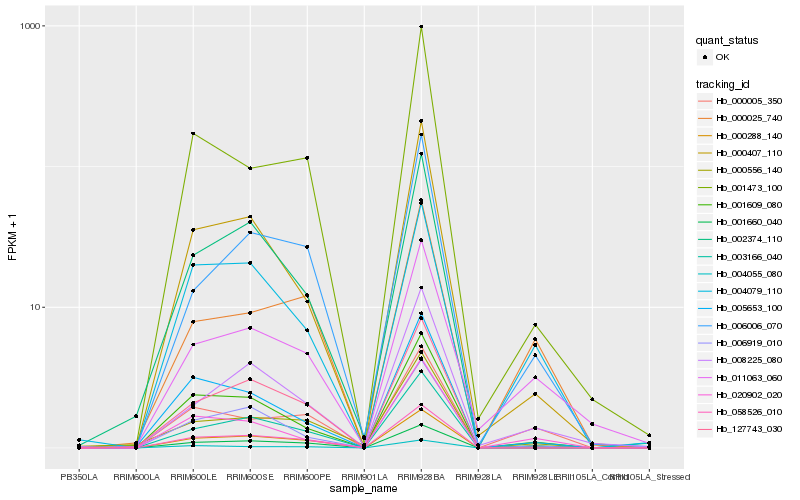
Hb_000288_140 |
0.0 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 35 [Jatropha curcas] |
| 2 |
Hb_001660_040 |
0.0595307816 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 3 |
Hb_001609_080 |
0.0998166928 |
- |
- |
hypothetical protein POPTR_0017s05660g [Populus trichocarpa] |
| 4 |
Hb_058526_010 |
0.1058803761 |
- |
- |
- |
| 5 |
Hb_000005_350 |
0.1141752559 |
- |
- |
PREDICTED: uncharacterized protein LOC105634263 [Jatropha curcas] |
| 6 |
Hb_003166_040 |
0.1143237364 |
- |
- |
PREDICTED: probable indole-3-acetic acid-amido synthetase GH3.1 [Jatropha curcas] |
| 7 |
Hb_127743_030 |
0.11668606 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_006006_070 |
0.1191927516 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_006919_010 |
0.1231612799 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Vitis vinifera] |
| 10 |
Hb_011063_060 |
0.1245593187 |
- |
- |
PREDICTED: endochitinase-like [Jatropha curcas] |
| 11 |
Hb_000556_140 |
0.1249018056 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 12 |
Hb_004079_110 |
0.1254678358 |
- |
- |
PREDICTED: uncharacterized protein LOC100257802 [Vitis vinifera] |
| 13 |
Hb_005653_100 |
0.1294763665 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 14 |
Hb_000025_740 |
0.130154333 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000407_110 |
0.13195629 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 16 |
Hb_020902_020 |
0.1360403598 |
- |
- |
hypothetical protein JCGZ_07820 [Jatropha curcas] |
| 17 |
Hb_008225_080 |
0.1371260304 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 18 |
Hb_002374_110 |
0.1381091053 |
- |
- |
PREDICTED: uncharacterized protein LOC105638965 [Jatropha curcas] |
| 19 |
Hb_001473_100 |
0.1385432802 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 20 |
Hb_004055_080 |
0.1386878171 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |