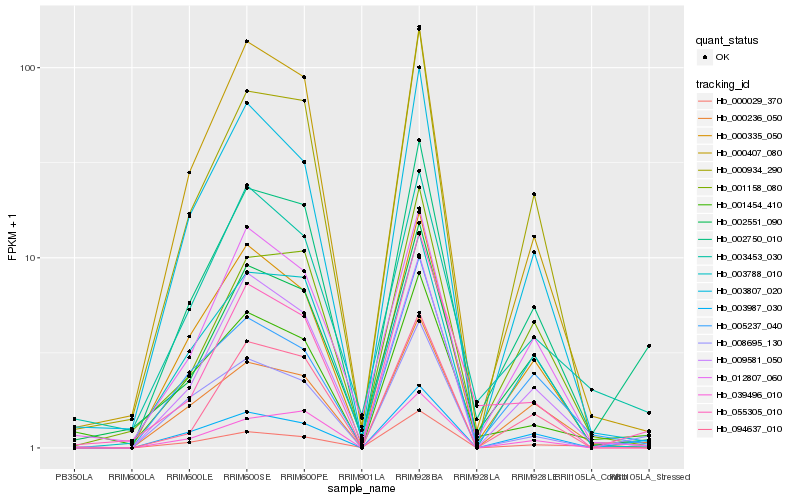
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002750_010 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 2 |
Hb_003987_030 |
0.1166187673 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 3 |
Hb_000236_050 |
0.1229421654 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 4 |
Hb_002551_090 |
0.1269968618 |
- |
- |
hypothetical protein JCGZ_17224 [Jatropha curcas] |
| 5 |
Hb_008695_130 |
0.1311432216 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000934_290 |
0.1337949534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009581_050 |
0.13427626 |
- |
- |
hypothetical protein CISIN_1g0025812mg, partial [Citrus sinensis] |
| 8 |
Hb_000335_050 |
0.1365239328 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003788_010 |
0.1405084717 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 10 |
Hb_055305_010 |
0.1412939916 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 11 |
Hb_001454_410 |
0.1444568125 |
- |
- |
PREDICTED: uncharacterized protein LOC105643453 [Jatropha curcas] |
| 12 |
Hb_003807_020 |
0.1463244434 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 13 |
Hb_003453_030 |
0.1474426684 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 14 |
Hb_000407_080 |
0.1474934442 |
- |
- |
1-aminocyclopropane-1-carboxylate oxidase [Hevea brasiliensis] |
| 15 |
Hb_001158_080 |
0.1480456445 |
- |
- |
PREDICTED: omega-hydroxypalmitate O-feruloyl transferase [Jatropha curcas] |
| 16 |
Hb_039496_010 |
0.1487314363 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_012807_060 |
0.1513706074 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_005237_040 |
0.152078122 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 19 |
Hb_000029_370 |
0.1522417835 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 20 |
Hb_094637_010 |
0.1523610946 |
- |
- |
PREDICTED: uncharacterized protein LOC105642042 [Jatropha curcas] |