| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
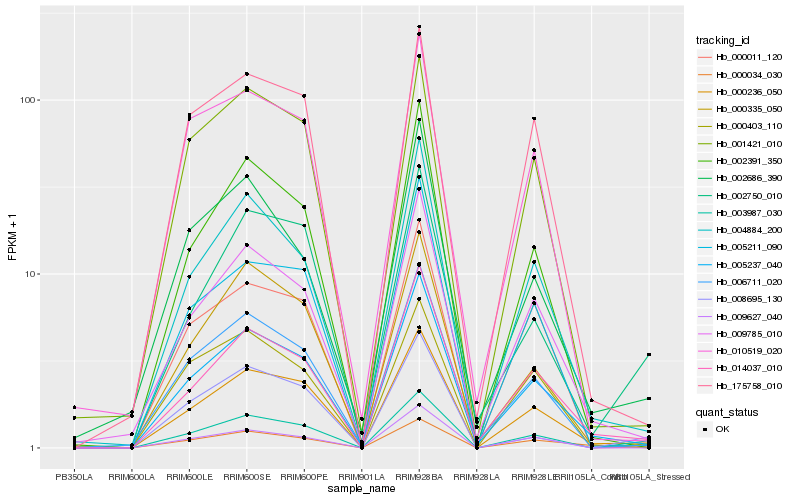
Hb_003987_030 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 2 |
Hb_000236_050 |
0.116272828 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 3 |
Hb_002750_010 |
0.1166187673 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 15 family protein [Populus trichocarpa] |
| 4 |
Hb_008695_130 |
0.1314063724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000034_030 |
0.135722382 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 6 |
Hb_006711_020 |
0.1425549562 |
- |
- |
hypothetical protein JCGZ_07258 [Jatropha curcas] |
| 7 |
Hb_005237_040 |
0.1481957535 |
- |
- |
PREDICTED: uncharacterized protein LOC105630923 [Jatropha curcas] |
| 8 |
Hb_014037_010 |
0.1510599336 |
- |
- |
PREDICTED: uncharacterized protein LOC105641871 [Jatropha curcas] |
| 9 |
Hb_004884_200 |
0.1511984002 |
- |
- |
PREDICTED: ferritin-3, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000403_110 |
0.1523705217 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 11 |
Hb_002391_350 |
0.1537236539 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 12 |
Hb_005211_090 |
0.1548973039 |
- |
- |
PREDICTED: aluminum-activated malate transporter 4-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_009785_010 |
0.1584230565 |
- |
- |
PREDICTED: uncharacterized protein LOC105648749 [Jatropha curcas] |
| 14 |
Hb_000011_120 |
0.1617668848 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 15 |
Hb_001421_010 |
0.1619099438 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009627_040 |
0.1625873188 |
- |
- |
Lipase class 3-related protein, putative [Theobroma cacao] |
| 17 |
Hb_175758_010 |
0.1637255697 |
- |
- |
PREDICTED: aspartic proteinase A1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_002686_390 |
0.1640093307 |
- |
- |
PREDICTED: uncharacterized protein LOC105647789 [Jatropha curcas] |
| 19 |
Hb_010519_020 |
0.1646980418 |
- |
- |
hypothetical protein JCGZ_19028 [Jatropha curcas] |
| 20 |
Hb_000335_050 |
0.1656560001 |
- |
- |
PREDICTED: CASP-like protein 4A1 isoform X1 [Jatropha curcas] |