| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
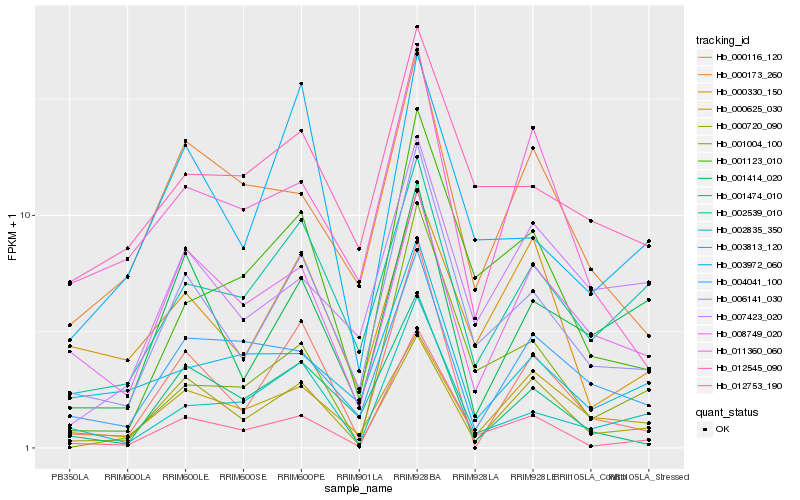
Hb_006141_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_012753_190 |
0.1511097063 |
- |
- |
PREDICTED: protein HAPLESS 2 [Populus euphratica] |
| 3 |
Hb_012545_090 |
0.1595401412 |
- |
- |
hypothetical protein JCGZ_04320 [Jatropha curcas] |
| 4 |
Hb_002835_350 |
0.1606369156 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 5 |
Hb_001123_010 |
0.164546395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002539_010 |
0.1697843029 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 7 |
Hb_000116_120 |
0.1728904072 |
- |
- |
PREDICTED: disease resistance response protein 206-like [Jatropha curcas] |
| 8 |
Hb_001474_010 |
0.1738456784 |
- |
- |
PREDICTED: synaptotagmin-3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000625_030 |
0.1774175378 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 10 |
Hb_000173_260 |
0.1782211794 |
- |
- |
hypothetical protein L484_010046 [Morus notabilis] |
| 11 |
Hb_003972_060 |
0.178713172 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 12 |
Hb_000720_090 |
0.1833177279 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 13 |
Hb_001414_020 |
0.1845451735 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_007423_020 |
0.1846166624 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 15 |
Hb_011360_060 |
0.1874658339 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 16 |
Hb_001004_100 |
0.1876424856 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_004041_100 |
0.188409065 |
- |
- |
- |
| 18 |
Hb_003813_120 |
0.1901510652 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_008749_020 |
0.1903063625 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000330_150 |
0.1923870594 |
- |
- |
PREDICTED: dihydroflavonol-4-reductase-like [Jatropha curcas] |