| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
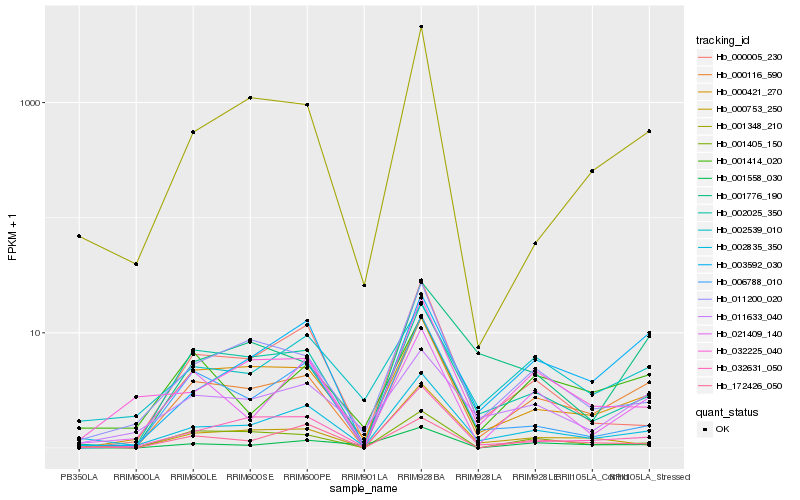
Hb_000116_590 |
0.0 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 2 |
Hb_000421_270 |
0.1351664223 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 3 |
Hb_002025_350 |
0.1505616465 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 4 |
Hb_002835_350 |
0.1700208848 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 5 |
Hb_001405_150 |
0.1703678519 |
- |
- |
PREDICTED: protein At-4/1 [Jatropha curcas] |
| 6 |
Hb_032631_050 |
0.1765694515 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 7 |
Hb_003592_030 |
0.1807726397 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 8 |
Hb_011200_020 |
0.1831210762 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 9 |
Hb_021409_140 |
0.1902751611 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 10 |
Hb_001348_210 |
0.1964923776 |
- |
- |
defensin protein precursor [Capsicum annuum] |
| 11 |
Hb_011633_040 |
0.2031861834 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 12 |
Hb_001414_020 |
0.2036417785 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000753_250 |
0.2061620908 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 14 |
Hb_006788_010 |
0.2110081754 |
- |
- |
PREDICTED: calcium-transporting ATPase 12, plasma membrane-type [Jatropha curcas] |
| 15 |
Hb_172426_050 |
0.2115994493 |
- |
- |
PREDICTED: uncharacterized protein LOC105634266 [Jatropha curcas] |
| 16 |
Hb_001776_190 |
0.2131694364 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_032225_040 |
0.2144416612 |
- |
- |
- |
| 18 |
Hb_000005_230 |
0.2152638669 |
- |
- |
hypothetical protein POPTR_0009s01160g [Populus trichocarpa] |
| 19 |
Hb_001558_030 |
0.2153300345 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 20 |
Hb_002539_010 |
0.2171944978 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |