| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
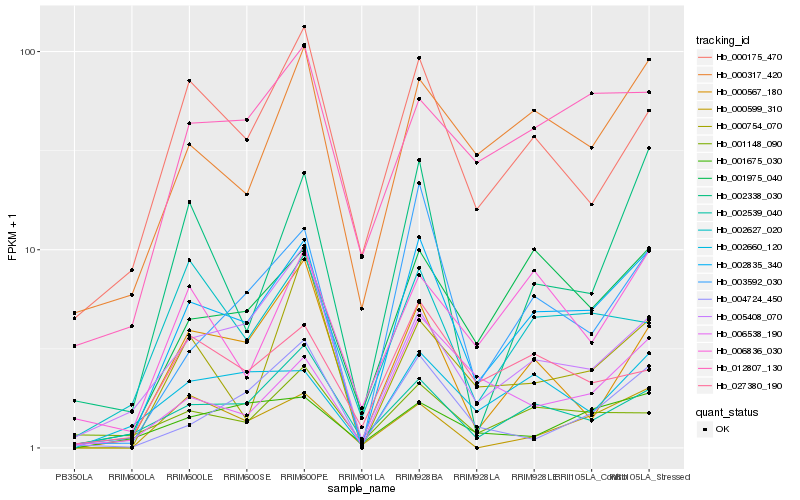
Hb_002835_340 |
0.0 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 2 |
Hb_001975_040 |
0.1582746946 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 3 |
Hb_000317_420 |
0.1597588341 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 4 |
Hb_003592_030 |
0.1668885184 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 5 |
Hb_002539_040 |
0.1813533782 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_012807_130 |
0.1876370764 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 7 |
Hb_002660_120 |
0.1885287235 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 8 |
Hb_005408_070 |
0.1900436108 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 9 |
Hb_002338_030 |
0.1919476173 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1-like [Tarenaya hassleriana] |
| 10 |
Hb_006836_030 |
0.1929268902 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 11 |
Hb_006538_190 |
0.1972072088 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000175_470 |
0.1977596523 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 13 |
Hb_027380_190 |
0.19863069 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000567_180 |
0.2007477478 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 15 |
Hb_002627_020 |
0.2040014506 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 16 |
Hb_001675_030 |
0.2086974917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004724_450 |
0.2088283946 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001148_090 |
0.209653328 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 19 |
Hb_000599_310 |
0.2110070632 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 20 |
Hb_000754_070 |
0.2162240714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |