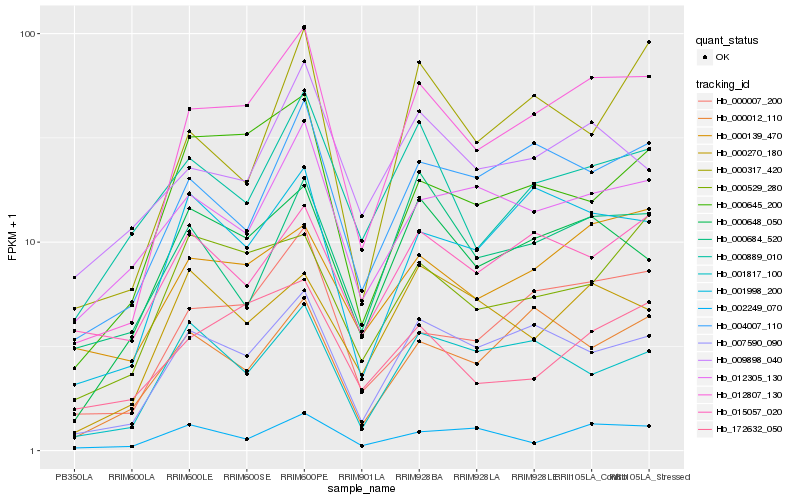
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012807_130 |
0.0 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 2 |
Hb_000007_200 |
0.1042335453 |
- |
- |
PREDICTED: choline/ethanolaminephosphotransferase 1 [Jatropha curcas] |
| 3 |
Hb_007590_090 |
0.1208308439 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 4 |
Hb_004007_110 |
0.1347472205 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 5 |
Hb_000648_050 |
0.1376541436 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001998_200 |
0.1405469985 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 7 |
Hb_000645_200 |
0.1448470015 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 8 |
Hb_000139_470 |
0.1494102182 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 9 |
Hb_000012_110 |
0.1531875317 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 10 |
Hb_172632_050 |
0.1573710293 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 11 |
Hb_000889_010 |
0.1578421054 |
transcription factor |
TF Family: MYB-related |
hypothetical protein PRUPE_ppa014728mg [Prunus persica] |
| 12 |
Hb_000317_420 |
0.1613032293 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 13 |
Hb_012305_130 |
0.1621397042 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 14 |
Hb_000529_280 |
0.1624252189 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001817_100 |
0.1634280877 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 16 |
Hb_009898_040 |
0.1637338178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000684_520 |
0.1682168836 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 18 |
Hb_002249_070 |
0.1694727868 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 19 |
Hb_000270_180 |
0.1705699621 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 20 |
Hb_015057_020 |
0.1706816367 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |