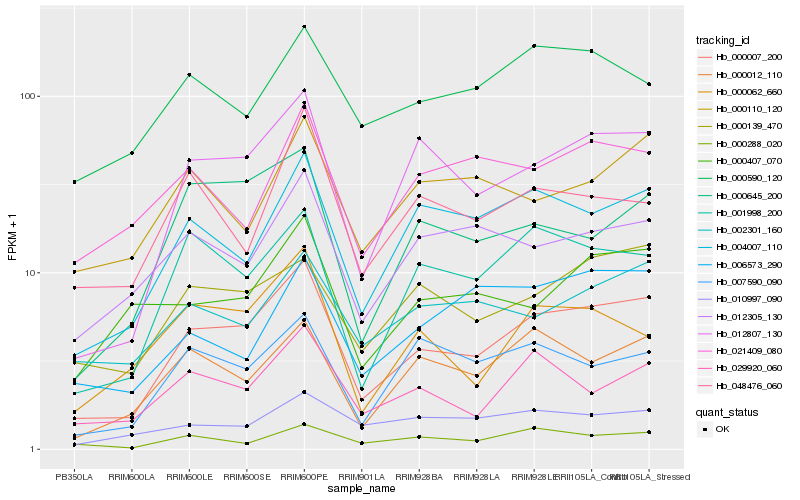
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000007_200 |
0.0 |
- |
- |
PREDICTED: choline/ethanolaminephosphotransferase 1 [Jatropha curcas] |
| 2 |
Hb_012807_130 |
0.1042335453 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 3 |
Hb_004007_110 |
0.1308310084 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 4 |
Hb_001998_200 |
0.1344346092 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 5 |
Hb_000012_110 |
0.1416273407 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 6 |
Hb_029920_060 |
0.1439274298 |
- |
- |
PREDICTED: UPF0554 protein-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_000139_470 |
0.147313721 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 8 |
Hb_000645_200 |
0.1482707654 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 9 |
Hb_012305_130 |
0.1521745933 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 10 |
Hb_000062_660 |
0.1541821277 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 11 |
Hb_007590_090 |
0.1549546118 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 12 |
Hb_002301_160 |
0.1576354588 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 13 |
Hb_000407_070 |
0.1590255891 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000288_020 |
0.1593397829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000590_120 |
0.1643772783 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 16 |
Hb_010997_090 |
0.1644623791 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 17 |
Hb_048476_060 |
0.1654486303 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000110_120 |
0.1655229278 |
- |
- |
PREDICTED: ras-related protein RABE1a [Fragaria vesca subsp. vesca] |
| 19 |
Hb_021409_080 |
0.1665064193 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 20 |
Hb_006573_290 |
0.1669790235 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |