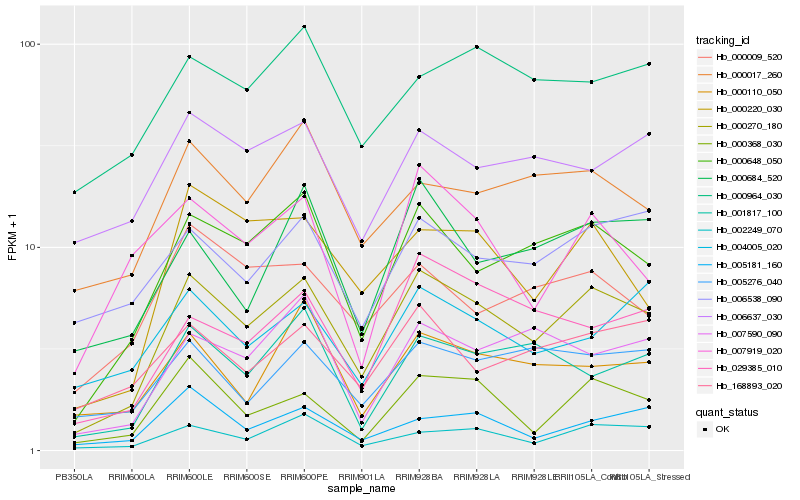
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_180 |
0.0 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 2 |
Hb_000648_050 |
0.103612774 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002249_070 |
0.1383322796 |
- |
- |
gcn4-complementing protein, putative [Ricinus communis] |
| 4 |
Hb_000368_030 |
0.1395596295 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 5 |
Hb_000220_030 |
0.1434499457 |
- |
- |
PREDICTED: amino-acid permease BAT1 homolog isoform X2 [Jatropha curcas] |
| 6 |
Hb_000684_520 |
0.1462065487 |
- |
- |
glutathione S-transferase L3-like [Jatropha curcas] |
| 7 |
Hb_029385_010 |
0.1478522428 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 8 |
Hb_168893_020 |
0.1516139947 |
- |
- |
PREDICTED: probable protein S-acyltransferase 15 [Jatropha curcas] |
| 9 |
Hb_007590_090 |
0.1518008853 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 10 |
Hb_001817_100 |
0.1524400649 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 11 |
Hb_005276_040 |
0.1535187289 |
transcription factor |
TF Family: TRAF |
PREDICTED: ARM REPEAT PROTEIN INTERACTING WITH ABF2-like [Jatropha curcas] |
| 12 |
Hb_006538_090 |
0.1546956244 |
- |
- |
PREDICTED: IST1 homolog [Jatropha curcas] |
| 13 |
Hb_000017_260 |
0.1552201586 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 14 |
Hb_000009_520 |
0.1584345029 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 15 |
Hb_000964_030 |
0.1590689099 |
- |
- |
ADP/ATP carrier 2 [Theobroma cacao] |
| 16 |
Hb_000110_050 |
0.1593225487 |
- |
- |
- |
| 17 |
Hb_006637_030 |
0.1606871161 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 18 |
Hb_004005_020 |
0.161913785 |
- |
- |
PREDICTED: nudix hydrolase 20, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_005181_160 |
0.1620999547 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 20 |
Hb_007919_020 |
0.1635112434 |
- |
- |
electron transporter, putative [Ricinus communis] |