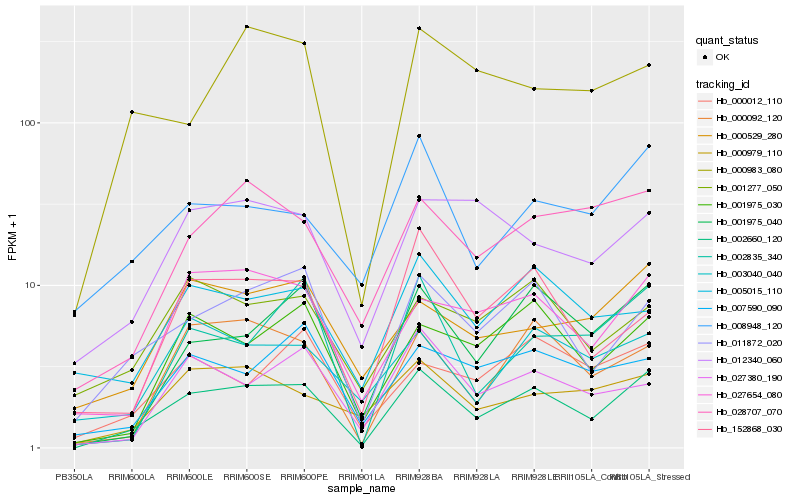
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002660_120 |
0.0 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 2 |
Hb_011872_020 |
0.1426003866 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 3 |
Hb_001975_040 |
0.1645794115 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 4 |
Hb_000092_120 |
0.1653265966 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 5 |
Hb_027654_080 |
0.1732585265 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_001975_030 |
0.1751275919 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 7 |
Hb_028707_070 |
0.1842114524 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_008948_120 |
0.1874404537 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_152868_030 |
0.1883549076 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 10 |
Hb_002835_340 |
0.1885287235 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 11 |
Hb_000529_280 |
0.1886936886 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000979_110 |
0.1919231124 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 13 |
Hb_027380_190 |
0.1947342103 |
- |
- |
PREDICTED: uncharacterized protein LOC105634028 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003040_040 |
0.1951143787 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_007590_090 |
0.1963017106 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 16 |
Hb_000983_080 |
0.196638833 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 17 |
Hb_001277_050 |
0.1968133371 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 18 |
Hb_012340_060 |
0.1991417286 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000012_110 |
0.2001207894 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 20 |
Hb_005015_110 |
0.2003166077 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |