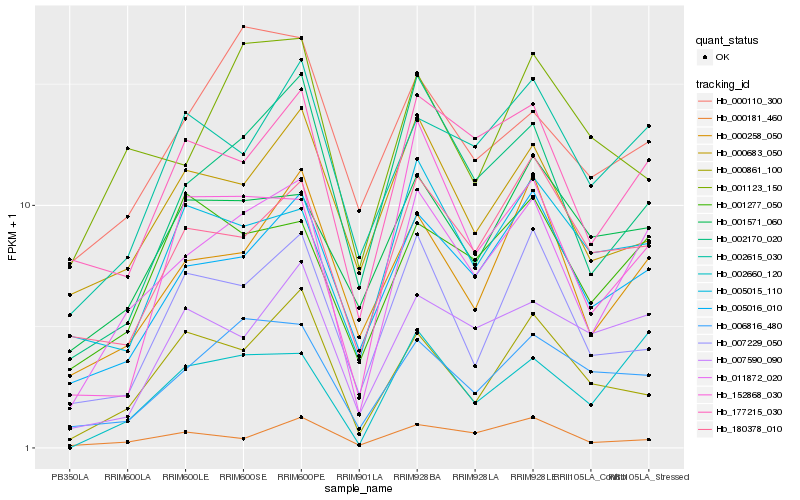
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011872_020 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 2 |
Hb_005016_010 |
0.1297236227 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 3 |
Hb_177215_030 |
0.1372609589 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_002170_020 |
0.1389626584 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 5 |
Hb_000258_050 |
0.1409830938 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002660_120 |
0.1426003866 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 7 |
Hb_006816_480 |
0.1490690759 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002615_030 |
0.151229473 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 9 |
Hb_180378_010 |
0.1579196743 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Populus euphratica] |
| 10 |
Hb_001277_050 |
0.1637722497 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000683_050 |
0.1643319671 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 12 |
Hb_007590_090 |
0.1649211288 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 13 |
Hb_001571_060 |
0.167609049 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 14 |
Hb_000181_460 |
0.1696895583 |
- |
- |
cmp-2-keto-3-deoctulosonate (cmp-kdo) cytidyltransferase, putative [Ricinus communis] |
| 15 |
Hb_000861_100 |
0.1699636632 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_007229_050 |
0.1706720293 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005015_110 |
0.1716572413 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 18 |
Hb_001123_150 |
0.1729699962 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 19 |
Hb_152868_030 |
0.1730141398 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 20 |
Hb_000110_300 |
0.1737098462 |
- |
- |
PREDICTED: uncharacterized protein LOC105642013 [Jatropha curcas] |