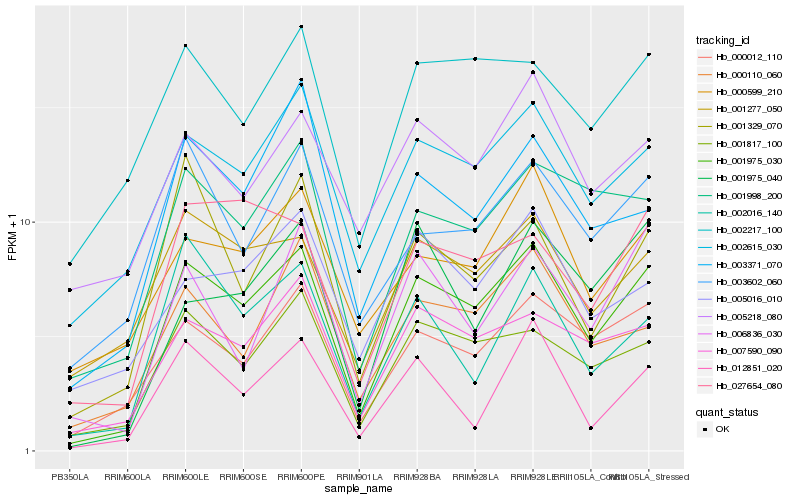
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001975_030 |
0.0 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 2 |
Hb_001817_100 |
0.1152361465 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 3 |
Hb_007590_090 |
0.1165254995 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 4 |
Hb_000012_110 |
0.1198176311 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 5 |
Hb_002615_030 |
0.1214953605 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 6 |
Hb_001998_200 |
0.1319334385 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 7 |
Hb_000110_060 |
0.1414745574 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 8 |
Hb_001277_050 |
0.1437912931 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 9 |
Hb_003602_060 |
0.1445616326 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 10 |
Hb_027654_080 |
0.1458162119 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002217_100 |
0.1468110266 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
dihydrolipoamide dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000599_210 |
0.147272758 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 13 |
Hb_003371_070 |
0.1487327809 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_001975_040 |
0.1508509971 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 15 |
Hb_006836_030 |
0.1511634328 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 16 |
Hb_005218_080 |
0.1541091928 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_012851_020 |
0.1552135443 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_001329_070 |
0.1553944008 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 19 |
Hb_002016_140 |
0.1554389574 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 20 |
Hb_005016_010 |
0.1563918622 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |