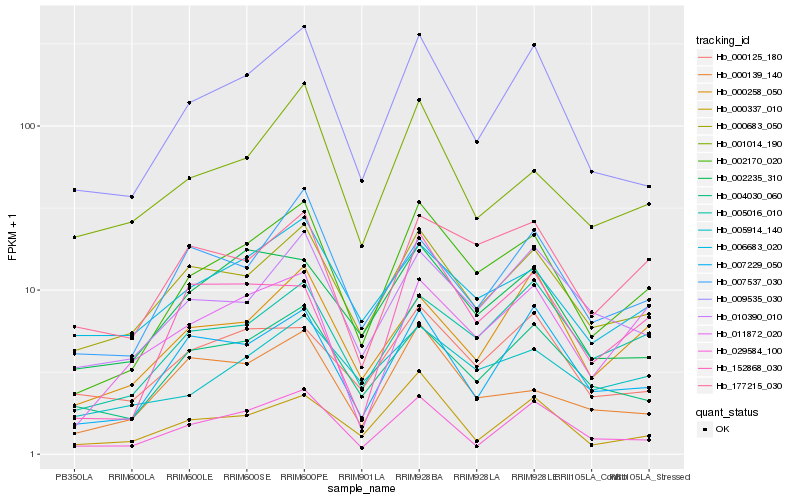
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002170_020 |
0.0 |
transcription factor |
TF Family: MYB |
MYB transcription factor [Hevea brasiliensis] |
| 2 |
Hb_009535_030 |
0.1194536282 |
- |
- |
CP2 [Hevea brasiliensis] |
| 3 |
Hb_005016_010 |
0.1251709157 |
- |
- |
AMP dependent CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_000683_050 |
0.1253215887 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 5 |
Hb_004030_060 |
0.134933917 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 6 |
Hb_006683_020 |
0.1380452992 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 7 |
Hb_011872_020 |
0.1389626584 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH128 [Jatropha curcas] |
| 8 |
Hb_177215_030 |
0.1402951472 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_005914_140 |
0.1414341619 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 10 |
Hb_010390_010 |
0.1418429119 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 11 |
Hb_007229_050 |
0.1422423528 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000258_050 |
0.1469165193 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002235_310 |
0.1476521124 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 14 |
Hb_001014_190 |
0.1496965024 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 15 |
Hb_007537_030 |
0.1497711015 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_029584_100 |
0.1500247172 |
- |
- |
Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase [Gossypium arboreum] |
| 17 |
Hb_152868_030 |
0.1514294801 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 18 |
Hb_000139_140 |
0.1517503239 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 19 |
Hb_000125_180 |
0.1536390921 |
- |
- |
PREDICTED: flowering time control protein FPA isoform X1 [Jatropha curcas] |
| 20 |
Hb_000337_010 |
0.1539749591 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |