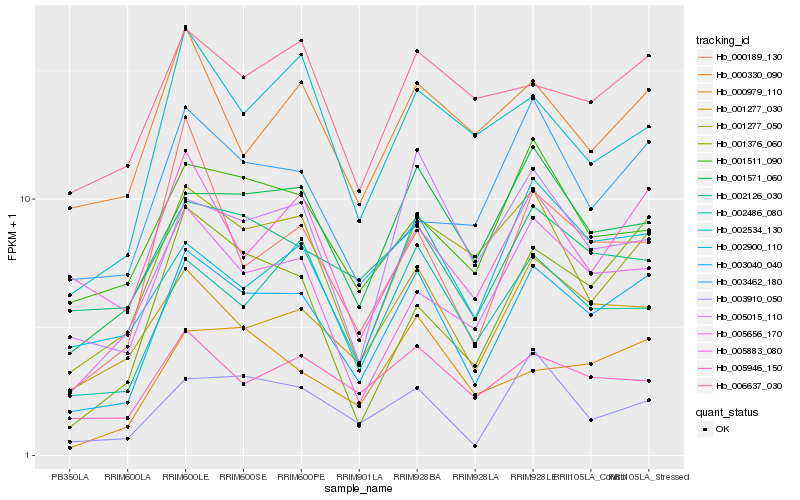
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003040_040 |
0.0 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001277_030 |
0.1189565226 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Nelumbo nucifera] |
| 3 |
Hb_001571_060 |
0.1222979164 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 4 |
Hb_002486_080 |
0.12890715 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 5 |
Hb_001277_050 |
0.1308991876 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005656_170 |
0.1349265035 |
- |
- |
PREDICTED: uncharacterized protein LOC105637447 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005946_150 |
0.1382831246 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 8 |
Hb_005015_110 |
0.1386520974 |
- |
- |
PREDICTED: 4-alpha-glucanotransferase, chloroplastic/amyloplastic [Jatropha curcas] |
| 9 |
Hb_000979_110 |
0.1392436906 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 10 |
Hb_003910_050 |
0.1400824196 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 11 |
Hb_002900_110 |
0.1424124217 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_001376_060 |
0.1426124411 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC isoform X2 [Jatropha curcas] |
| 13 |
Hb_005883_080 |
0.1457291262 |
- |
- |
type 2A protein phosphatase-2 [Populus trichocarpa] |
| 14 |
Hb_001511_090 |
0.1467974176 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 15 |
Hb_006637_030 |
0.1477623213 |
- |
- |
hypothetical protein CISIN_1g017413mg [Citrus sinensis] |
| 16 |
Hb_002126_030 |
0.1489713817 |
- |
- |
ferrochelatase, putative [Ricinus communis] |
| 17 |
Hb_003462_180 |
0.1491467195 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 18 |
Hb_002534_130 |
0.1494765257 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 19 |
Hb_000189_130 |
0.1496073559 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000330_090 |
0.1497298231 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |