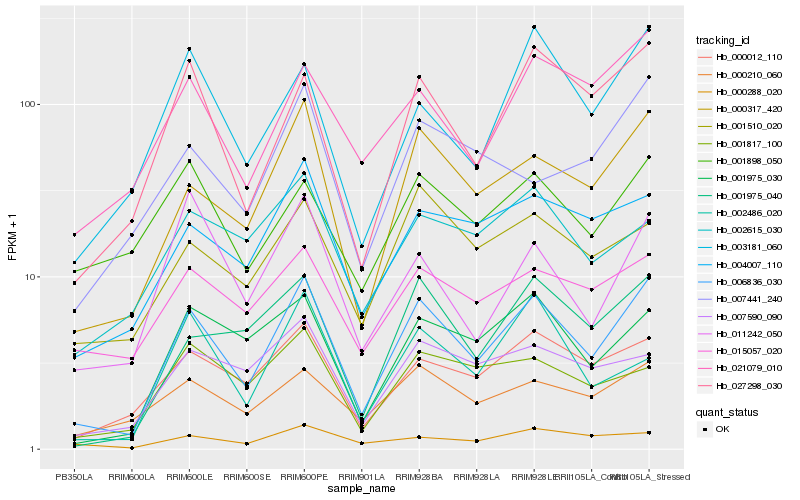
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006836_030 |
0.0 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 2 |
Hb_000317_420 |
0.1097887847 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 3 |
Hb_027298_030 |
0.1356569068 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 4 |
Hb_000012_110 |
0.1454994033 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 5 |
Hb_001975_040 |
0.148475551 |
rubber biosynthesis |
Gene Name: 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase |
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase [Hevea brasiliensis] |
| 6 |
Hb_001975_030 |
0.1511634328 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 7 |
Hb_004007_110 |
0.1554894666 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 8 |
Hb_000210_060 |
0.158115306 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 9 |
Hb_021079_010 |
0.1605641559 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 10 |
Hb_003181_060 |
0.1635115978 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 11 |
Hb_001510_020 |
0.1652581361 |
- |
- |
Diaminopimelate epimerase, putative [Ricinus communis] |
| 12 |
Hb_007590_090 |
0.1683450072 |
- |
- |
PREDICTED: O-glucosyltransferase rumi [Jatropha curcas] |
| 13 |
Hb_011242_050 |
0.1729568667 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 14 |
Hb_015057_020 |
0.1730759638 |
- |
- |
PREDICTED: E3 SUMO-protein ligase MMS21 [Jatropha curcas] |
| 15 |
Hb_001817_100 |
0.1733043061 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 16 |
Hb_000288_020 |
0.1742044832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002486_020 |
0.1757193779 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_007441_240 |
0.1804838431 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 19 |
Hb_001898_050 |
0.180997279 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 20 |
Hb_002615_030 |
0.1813325411 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |