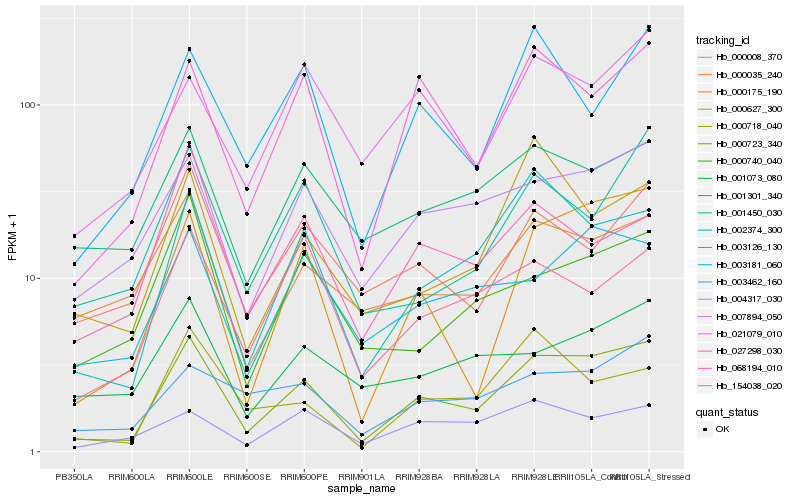
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_340 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 2 |
Hb_000175_190 |
0.1410624572 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 3 |
Hb_027298_030 |
0.1415338338 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 4 |
Hb_002374_300 |
0.1417645356 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 5 |
Hb_007894_050 |
0.1493630984 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003126_130 |
0.1651400982 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 7 |
Hb_003181_060 |
0.165847532 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 8 |
Hb_001301_340 |
0.1728224603 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 9 |
Hb_000035_240 |
0.1731465984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000008_370 |
0.1737001499 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |
| 11 |
Hb_003462_160 |
0.1744074578 |
transcription factor |
TF Family: GNAT |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 12 |
Hb_001450_030 |
0.1750110771 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 13 |
Hb_154038_020 |
0.1757440331 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 14 |
Hb_021079_010 |
0.1775830384 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 15 |
Hb_004317_030 |
0.1788394304 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 16 |
Hb_000740_040 |
0.1791142277 |
- |
- |
PREDICTED: uncharacterized protein LOC105126082 [Populus euphratica] |
| 17 |
Hb_068194_010 |
0.179357015 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 18 |
Hb_000718_040 |
0.1813811559 |
- |
- |
PREDICTED: lipase member N [Jatropha curcas] |
| 19 |
Hb_001073_080 |
0.1817432906 |
- |
- |
PREDICTED: protein root UVB sensitive 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000627_300 |
0.1823035914 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |