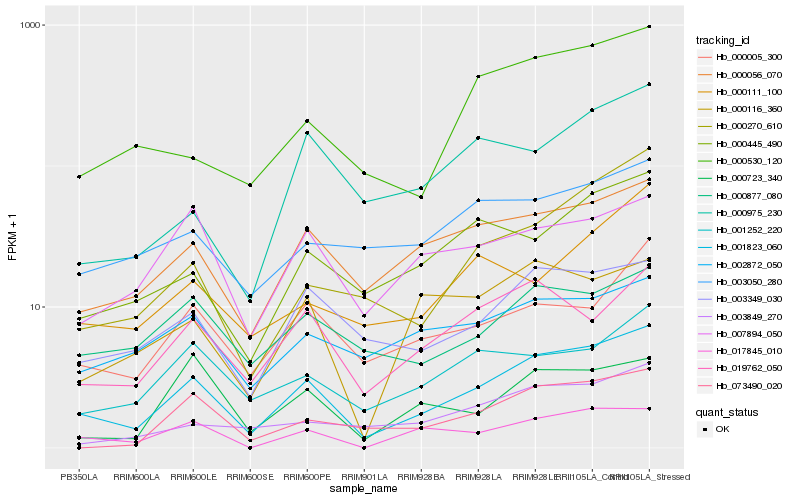
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001823_060 |
0.0 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 2 |
Hb_000056_070 |
0.1511167609 |
- |
- |
- |
| 3 |
Hb_003349_030 |
0.1570981834 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 4 |
Hb_019762_050 |
0.1590826511 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 5 |
Hb_000005_300 |
0.1652219366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_017845_010 |
0.1667262558 |
- |
- |
- |
| 7 |
Hb_001252_220 |
0.1704264167 |
- |
- |
PREDICTED: uncharacterized protein LOC105640647 [Jatropha curcas] |
| 8 |
Hb_000445_490 |
0.1718984986 |
- |
- |
- |
| 9 |
Hb_000530_120 |
0.1720672542 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 10 |
Hb_000877_080 |
0.1754449236 |
- |
- |
PREDICTED: uncharacterized protein LOC105640901 [Jatropha curcas] |
| 11 |
Hb_000270_610 |
0.1754717721 |
- |
- |
PREDICTED: uncharacterized protein At5g50100, mitochondrial [Jatropha curcas] |
| 12 |
Hb_073490_020 |
0.1813220755 |
- |
- |
hypothetical protein PRUPE_ppa016333mg, partial [Prunus persica] |
| 13 |
Hb_007894_050 |
0.1831785642 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000116_360 |
0.1852372507 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 15 |
Hb_002872_050 |
0.1858840999 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003849_270 |
0.1865448265 |
- |
- |
- |
| 17 |
Hb_003050_280 |
0.1887876754 |
transcription factor |
TF Family: HMG |
DNA-binding protein MNB1B, putative [Ricinus communis] |
| 18 |
Hb_000111_100 |
0.1893308756 |
- |
- |
PREDICTED: NADPH:quinone oxidoreductase-like [Camelina sativa] |
| 19 |
Hb_000723_340 |
0.1902465784 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 20 |
Hb_000975_230 |
0.1902796483 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |