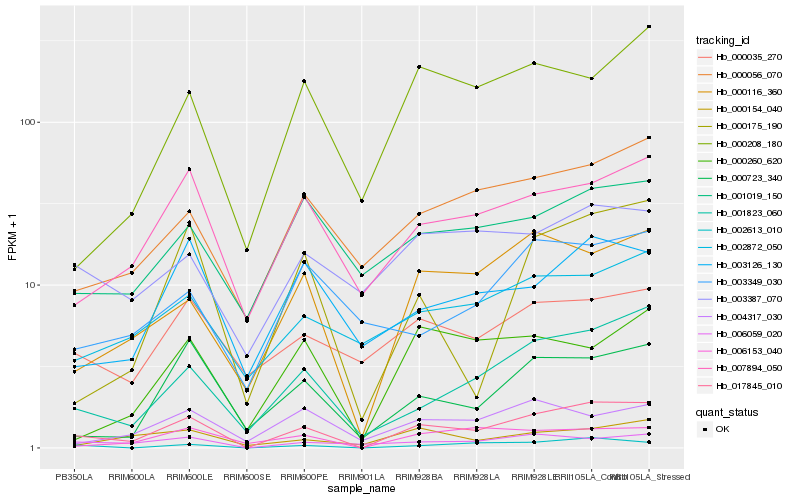
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017845_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_001823_060 |
0.1667262558 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 3 |
Hb_000116_360 |
0.1777468863 |
- |
- |
PREDICTED: uncharacterized protein LOC105116418 [Populus euphratica] |
| 4 |
Hb_002613_010 |
0.1887280176 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 5 |
Hb_000723_340 |
0.19556085 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 6 |
Hb_007894_050 |
0.1959443603 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000056_070 |
0.202474539 |
- |
- |
- |
| 8 |
Hb_000175_190 |
0.2105006152 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 9 |
Hb_006059_020 |
0.2115802793 |
- |
- |
PREDICTED: uncharacterized protein LOC105648672 [Jatropha curcas] |
| 10 |
Hb_006153_040 |
0.2141357909 |
- |
- |
hypothetical protein PRUPE_ppa014788mg [Prunus persica] |
| 11 |
Hb_002872_050 |
0.2161376142 |
- |
- |
PREDICTED: thioredoxin-like 3-2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000208_180 |
0.2169185159 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 13 |
Hb_003126_130 |
0.2172854222 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 14 |
Hb_000035_270 |
0.2218166405 |
- |
- |
PREDICTED: uncharacterized protein LOC105637542 [Jatropha curcas] |
| 15 |
Hb_000260_620 |
0.2223954043 |
- |
- |
Thermosensitive gluconokinase, putative [Ricinus communis] |
| 16 |
Hb_003387_070 |
0.2232599722 |
- |
- |
Acyl-protein thioesterase, putative [Ricinus communis] |
| 17 |
Hb_001019_150 |
0.2233139222 |
- |
- |
Acyl-protein thioesterase, putative [Ricinus communis] |
| 18 |
Hb_000154_040 |
0.2260276333 |
- |
- |
hypothetical protein JCGZ_25602 [Jatropha curcas] |
| 19 |
Hb_003349_030 |
0.2302879273 |
- |
- |
PREDICTED: uncharacterized protein LOC105628283 [Jatropha curcas] |
| 20 |
Hb_004317_030 |
0.2308040183 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |