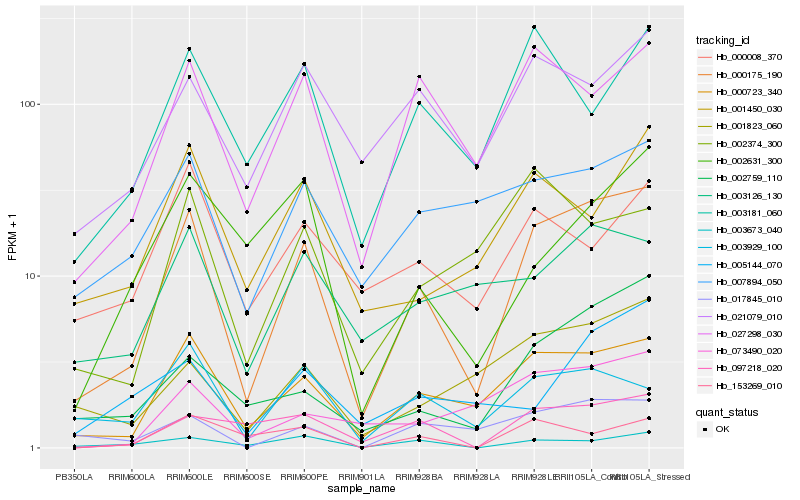
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_190 |
0.0 |
- |
- |
PREDICTED: phosphatidylcholine:diacylglycerol cholinephosphotransferase 1-like [Jatropha curcas] |
| 2 |
Hb_000723_340 |
0.1410624572 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 3 |
Hb_027298_030 |
0.1978223182 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 4 |
Hb_017845_010 |
0.2105006152 |
- |
- |
- |
| 5 |
Hb_002759_110 |
0.2116814141 |
- |
- |
RNA-binding family protein, putative isoform 1 [Theobroma cacao] |
| 6 |
Hb_001823_060 |
0.2117906023 |
- |
- |
fructose-bisphosphate aldolase, putative [Ricinus communis] |
| 7 |
Hb_097218_020 |
0.2150888473 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 8 |
Hb_021079_010 |
0.2153456967 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 9 |
Hb_003673_040 |
0.2172336533 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 10 |
Hb_003181_060 |
0.2225195322 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 11 |
Hb_001450_030 |
0.2245351054 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 12 |
Hb_007894_050 |
0.2266874952 |
- |
- |
PREDICTED: reactive Intermediate Deaminase A, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003929_100 |
0.2292366386 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Populus euphratica] |
| 14 |
Hb_073490_020 |
0.2303380299 |
- |
- |
hypothetical protein PRUPE_ppa016333mg, partial [Prunus persica] |
| 15 |
Hb_003126_130 |
0.2338581185 |
- |
- |
inositol-1-monophosphatase family protein [Populus trichocarpa] |
| 16 |
Hb_005144_070 |
0.2399235881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_153269_010 |
0.2408717226 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 18 |
Hb_002631_300 |
0.240873878 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 19 |
Hb_002374_300 |
0.2412964757 |
- |
- |
hypothetical protein JCGZ_12140 [Jatropha curcas] |
| 20 |
Hb_000008_370 |
0.2445859516 |
- |
- |
PREDICTED: uncharacterized protein LOC105628095 [Jatropha curcas] |