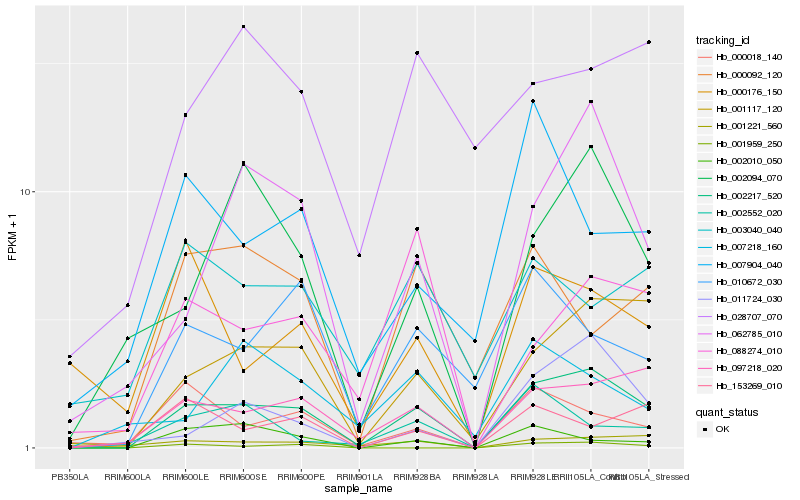
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_520 |
0.0 |
- |
- |
PREDICTED: probable protein phosphatase 2C 42 isoform X3 [Populus euphratica] |
| 2 |
Hb_001117_120 |
0.1808611673 |
- |
- |
chloride channel clc, putative [Ricinus communis] |
| 3 |
Hb_097218_020 |
0.1880345774 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 4 |
Hb_062785_010 |
0.2150776336 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 5 |
Hb_002094_070 |
0.2370691082 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 6 |
Hb_011724_030 |
0.2459027286 |
- |
- |
Organic cation transporter, putative [Ricinus communis] |
| 7 |
Hb_010672_030 |
0.2551771693 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 8 |
Hb_000018_140 |
0.2594053494 |
- |
- |
PREDICTED: uncharacterized protein LOC105638239 [Jatropha curcas] |
| 9 |
Hb_001959_250 |
0.2630611375 |
- |
- |
hypothetical protein CARUB_v10015435mg [Capsella rubella] |
| 10 |
Hb_088274_010 |
0.271280899 |
- |
- |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 11 |
Hb_153269_010 |
0.2743527237 |
- |
- |
hypothetical protein CICLE_v10029937mg [Citrus clementina] |
| 12 |
Hb_000092_120 |
0.2760230377 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 4 homolog A-like [Populus euphratica] |
| 13 |
Hb_002010_050 |
0.2765867463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002552_020 |
0.2807050952 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 15 |
Hb_028707_070 |
0.2820987752 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_000176_150 |
0.2826301766 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001221_560 |
0.2841630155 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase 1B-like [Jatropha curcas] |
| 18 |
Hb_003040_040 |
0.2865053587 |
- |
- |
PREDICTED: protein SAWADEE HOMEODOMAIN HOMOLOG 1-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_007218_160 |
0.2865079747 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 20 |
Hb_007904_040 |
0.2886828394 |
- |
- |
PREDICTED: uncharacterized protein LOC105644727 [Jatropha curcas] |