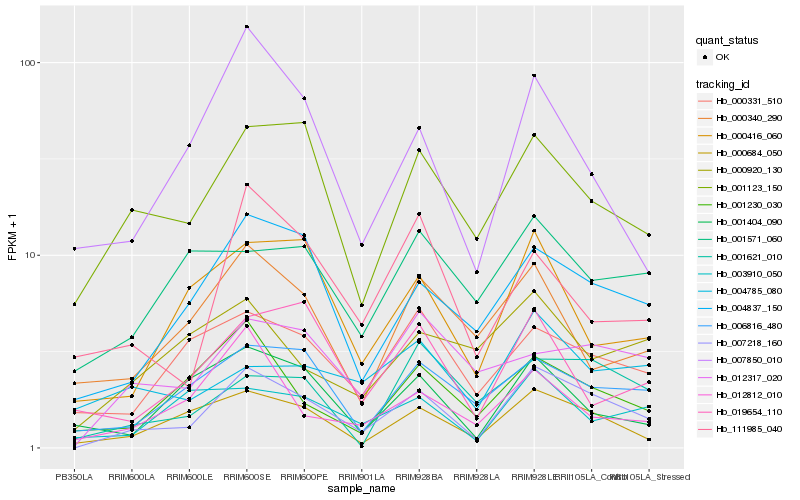
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007218_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 2 |
Hb_000684_050 |
0.1763404406 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |
| 3 |
Hb_004837_150 |
0.1829848492 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 4 |
Hb_001123_150 |
0.196675062 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 5 |
Hb_006816_480 |
0.2031966044 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001230_030 |
0.212118569 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |
| 7 |
Hb_000331_510 |
0.2131099479 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 8 |
Hb_000416_060 |
0.2141273115 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 9 |
Hb_012317_020 |
0.2149267427 |
- |
- |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 10 |
Hb_007850_010 |
0.221092085 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 11 |
Hb_003910_050 |
0.2309796344 |
- |
- |
PREDICTED: uncharacterized protein LOC105646926 [Jatropha curcas] |
| 12 |
Hb_000340_290 |
0.2314165302 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 13 |
Hb_001404_090 |
0.2317937162 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 14 |
Hb_111985_040 |
0.2324876415 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 3 [Jatropha curcas] |
| 15 |
Hb_001621_010 |
0.2326713083 |
- |
- |
PREDICTED: uncharacterized protein LOC102615432 [Citrus sinensis] |
| 16 |
Hb_012812_010 |
0.2342456475 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 17 |
Hb_001571_060 |
0.2344367366 |
- |
- |
PREDICTED: uncharacterized protein LOC105643976 [Jatropha curcas] |
| 18 |
Hb_004785_080 |
0.2351351385 |
- |
- |
hypothetical protein POPTR_0005s21250g [Populus trichocarpa] |
| 19 |
Hb_000920_130 |
0.2353287551 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_019654_110 |
0.235657794 |
- |
- |
PREDICTED: F-box protein SKIP23-like [Jatropha curcas] |