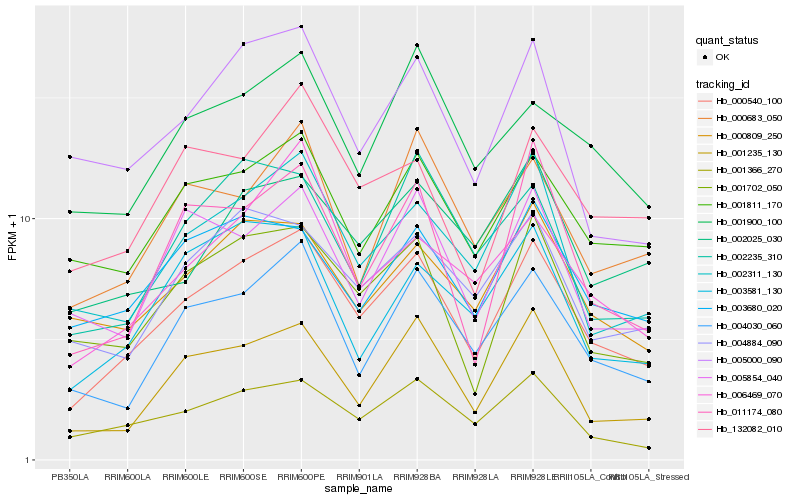
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000540_100 |
0.0 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 2 |
Hb_001366_270 |
0.0981577121 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 3 |
Hb_004030_060 |
0.1025385055 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 4 |
Hb_002311_130 |
0.103240285 |
- |
- |
PREDICTED: importin subunit alpha-2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001702_050 |
0.1039611222 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001235_130 |
0.1070723328 |
- |
- |
- |
| 7 |
Hb_001811_170 |
0.1087459214 |
- |
- |
dynamin, putative [Ricinus communis] |
| 8 |
Hb_004884_090 |
0.111010212 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 9 |
Hb_011174_080 |
0.1142086152 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 10 |
Hb_000809_250 |
0.1146338249 |
- |
- |
PREDICTED: uncharacterized protein LOC105639683 [Jatropha curcas] |
| 11 |
Hb_003680_020 |
0.1158158916 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_132082_010 |
0.1185337515 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 13 |
Hb_006469_070 |
0.1209684507 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g22040 [Jatropha curcas] |
| 14 |
Hb_001900_100 |
0.1216645605 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005000_090 |
0.1217442579 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |
| 16 |
Hb_000683_050 |
0.1217498027 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 17 |
Hb_002235_310 |
0.1225176571 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 18 |
Hb_005854_040 |
0.1226720113 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003581_130 |
0.1232980435 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_002025_030 |
0.1233045998 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |