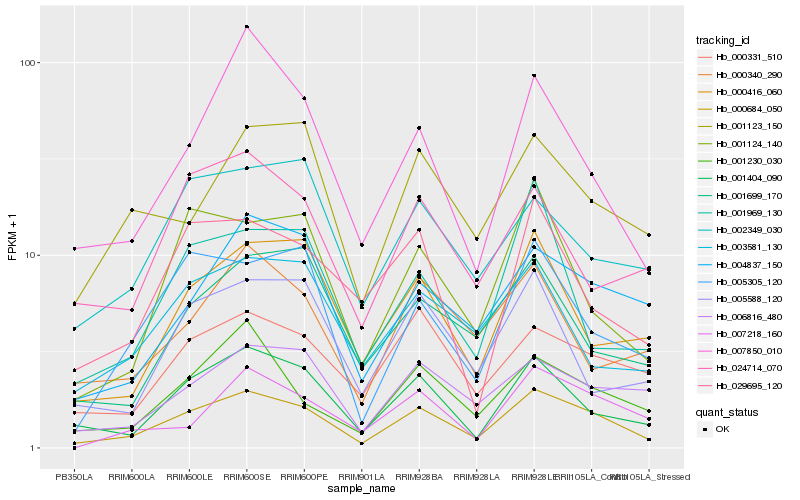
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_050 |
0.0 |
- |
- |
PREDICTED: MLO-like protein 6 [Sesamum indicum] |
| 2 |
Hb_001404_090 |
0.1350644249 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 3 |
Hb_001124_140 |
0.1589463531 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 4 |
Hb_005305_120 |
0.1593725385 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 5 |
Hb_001230_030 |
0.1597478726 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 isoform X2 [Populus euphratica] |
| 6 |
Hb_006816_480 |
0.1603419239 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000416_060 |
0.1609118761 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 8 |
Hb_000331_510 |
0.1611639359 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10-like [Jatropha curcas] |
| 9 |
Hb_004837_150 |
0.1628609021 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 10 |
Hb_007850_010 |
0.1640178416 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 11 |
Hb_001123_150 |
0.1688024807 |
transcription factor |
TF Family: NAC |
NAC transcription factor [Hevea brasiliensis] |
| 12 |
Hb_029695_120 |
0.1723586675 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 13 |
Hb_007218_160 |
0.1763404406 |
- |
- |
PREDICTED: uncharacterized protein LOC105108358 [Populus euphratica] |
| 14 |
Hb_003581_130 |
0.1770628827 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_005588_120 |
0.1775897657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001699_170 |
0.1785886092 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002349_030 |
0.1791974317 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_024714_070 |
0.1796712297 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 19 |
Hb_000340_290 |
0.1799233018 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog [Jatropha curcas] |
| 20 |
Hb_001969_130 |
0.1801202299 |
- |
- |
PREDICTED: molybdate transporter 2 [Jatropha curcas] |