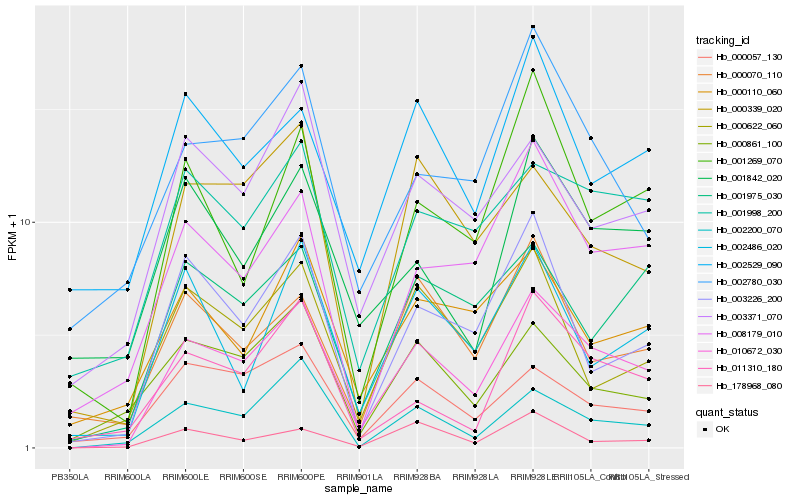
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010672_030 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 2 |
Hb_008179_010 |
0.1491282956 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 16 [Jatropha curcas] |
| 3 |
Hb_001998_200 |
0.1499416518 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 4 |
Hb_002200_070 |
0.1529809036 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 5 |
Hb_003226_200 |
0.1591476203 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 6 |
Hb_000057_130 |
0.1596314887 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 7 |
Hb_000622_060 |
0.160533235 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003371_070 |
0.1607295258 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001975_030 |
0.1627139346 |
- |
- |
4-diphosphocytidyl-2-c-methyl-d-erythritol kinase, partial [Plectranthus barbatus] |
| 10 |
Hb_001269_070 |
0.1632593261 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000339_020 |
0.164829581 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 12-like [Jatropha curcas] |
| 12 |
Hb_000110_060 |
0.1650304789 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 13 |
Hb_002780_030 |
0.1663167019 |
- |
- |
PREDICTED: molybdate transporter 1 [Jatropha curcas] |
| 14 |
Hb_000861_100 |
0.1670502797 |
- |
- |
PREDICTED: probable fucosyltransferase 8 isoform X1 [Jatropha curcas] |
| 15 |
Hb_011310_180 |
0.1676231258 |
- |
- |
PREDICTED: phospholipase D alpha 1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000070_110 |
0.1698501952 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002486_020 |
0.172223493 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002529_090 |
0.1724220218 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase [Hevea brasiliensis] |
| 19 |
Hb_001842_020 |
0.1729181779 |
- |
- |
PREDICTED: low molecular weight phosphotyrosine protein phosphatase [Jatropha curcas] |
| 20 |
Hb_178968_080 |
0.173596533 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X2 [Jatropha curcas] |