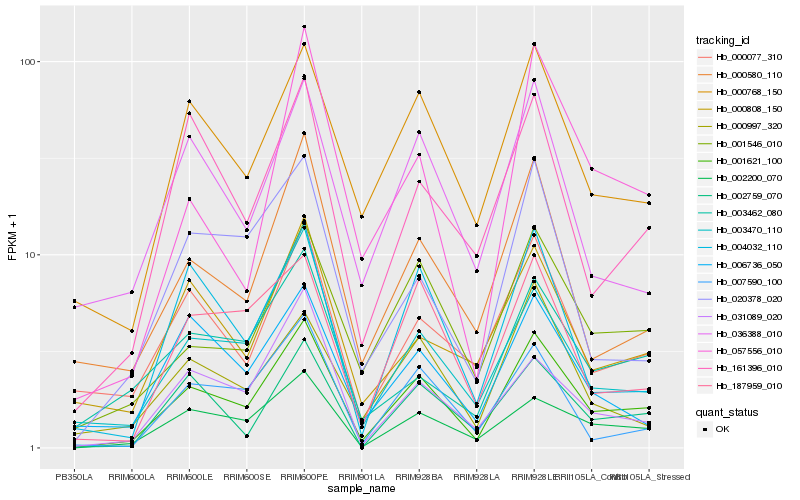
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001621_100 |
0.0 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4 [Jatropha curcas] |
| 2 |
Hb_002200_070 |
0.1255784132 |
- |
- |
PREDICTED: endoglucanase 9-like [Populus euphratica] |
| 3 |
Hb_000768_150 |
0.1396069886 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 4 |
Hb_001546_010 |
0.1438928774 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003470_110 |
0.1468397285 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000580_110 |
0.1553223163 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3 [Jatropha curcas] |
| 7 |
Hb_002759_070 |
0.1555069153 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 8 |
Hb_187959_010 |
0.1558745389 |
- |
- |
hypothetical protein JCGZ_12266 [Jatropha curcas] |
| 9 |
Hb_004032_110 |
0.1560897708 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000077_310 |
0.1562296881 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 11 |
Hb_007590_100 |
0.1592759105 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 12 |
Hb_161396_010 |
0.1620629687 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 13 |
Hb_031089_020 |
0.1665852093 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 14 |
Hb_000997_320 |
0.1668115833 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme-like [Malus domestica] |
| 15 |
Hb_036388_010 |
0.1672157712 |
- |
- |
PREDICTED: formate--tetrahydrofolate ligase [Populus euphratica] |
| 16 |
Hb_057556_010 |
0.1673210825 |
- |
- |
Light-regulated protein precursor, putative [Ricinus communis] |
| 17 |
Hb_000808_150 |
0.1687977298 |
- |
- |
PREDICTED: uncharacterized protein LOC105647444 [Jatropha curcas] |
| 18 |
Hb_003462_080 |
0.1690572879 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_020378_020 |
0.1692475623 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_006736_050 |
0.1695982201 |
- |
- |
hypothetical protein POPTR_0006s04450g [Populus trichocarpa] |