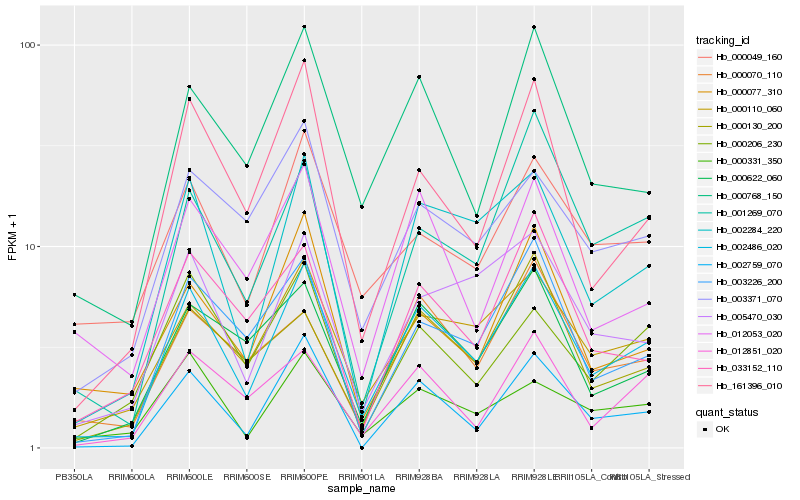
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002486_020 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_002284_220 |
0.1089736024 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 3 |
Hb_000110_060 |
0.1190258016 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 4 |
Hb_002759_070 |
0.1276002572 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 5 |
Hb_003226_200 |
0.1319063186 |
- |
- |
magnesium/proton exchanger, putative [Ricinus communis] |
| 6 |
Hb_000768_150 |
0.1350850358 |
- |
- |
PREDICTED: glycine-rich protein A3-like [Fragaria vesca subsp. vesca] |
| 7 |
Hb_005470_030 |
0.1356953019 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000622_060 |
0.1371140753 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_161396_010 |
0.1396364722 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 10 |
Hb_012851_020 |
0.1443138573 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_001269_070 |
0.1450494831 |
- |
- |
double-stranded RNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_000331_350 |
0.149430367 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 13 |
Hb_003371_070 |
0.1507429313 |
- |
- |
PREDICTED: citrate synthase, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000049_160 |
0.15229263 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 15 |
Hb_000130_200 |
0.1527272562 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X3 [Jatropha curcas] |
| 16 |
Hb_033152_110 |
0.1538021324 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 17 |
Hb_012053_020 |
0.1546163451 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000070_110 |
0.1556485578 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000077_310 |
0.1571856964 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 20 |
Hb_000206_230 |
0.1599324613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |