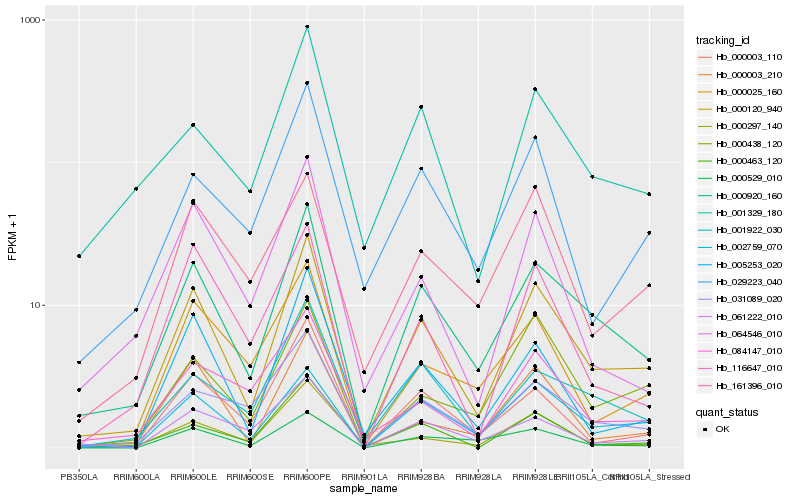
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_940 |
0.0 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000920_160 |
0.0869817859 |
- |
- |
annexin [Manihot esculenta] |
| 3 |
Hb_002759_070 |
0.1325959257 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 7 [Jatropha curcas] |
| 4 |
Hb_005253_020 |
0.1493091386 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 5 |
Hb_000463_120 |
0.1521704633 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 6 |
Hb_000003_210 |
0.152868129 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 7 |
Hb_000297_140 |
0.1533416179 |
- |
- |
hypothetical protein RCOM_0293450 [Ricinus communis] |
| 8 |
Hb_001922_030 |
0.1558358982 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 9 |
Hb_031089_020 |
0.1595945374 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 10 |
Hb_000438_120 |
0.1629047839 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 11 |
Hb_000529_010 |
0.1638511837 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 12 |
Hb_029223_040 |
0.1659137996 |
- |
- |
s-adenosylmethionine synthetase, putative [Ricinus communis] |
| 13 |
Hb_000003_110 |
0.1667433609 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_001329_180 |
0.1693954574 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 15 |
Hb_084147_010 |
0.1710154473 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 16 |
Hb_116647_010 |
0.1735657503 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000025_160 |
0.1737352132 |
- |
- |
PREDICTED: F-box protein At2g26850 isoform X1 [Jatropha curcas] |
| 18 |
Hb_064546_010 |
0.1745192334 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 19 |
Hb_161396_010 |
0.1748408606 |
- |
- |
hypothetical protein F383_26394 [Gossypium arboreum] |
| 20 |
Hb_061222_010 |
0.1750879089 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |