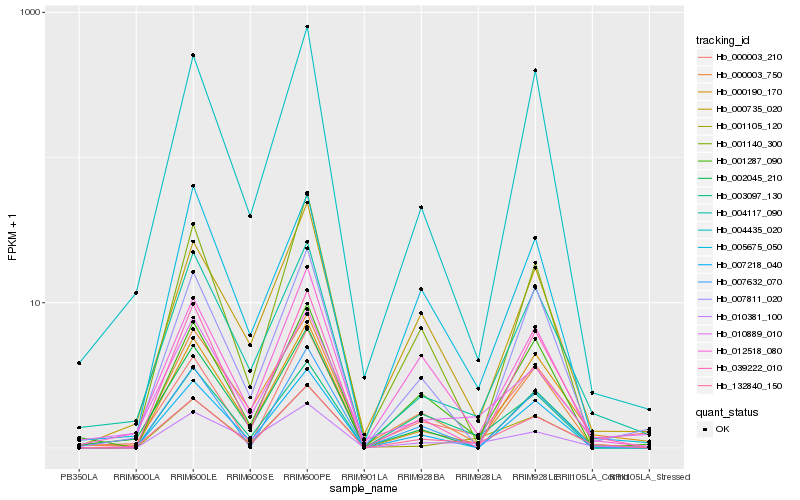
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132840_150 |
0.0 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 2 |
Hb_004117_090 |
0.1213064435 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 3 |
Hb_001105_120 |
0.1274353733 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 4 |
Hb_012518_080 |
0.1327711073 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 5 |
Hb_000190_170 |
0.1331827403 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 6 |
Hb_039222_010 |
0.1351882332 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000003_210 |
0.1364794613 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 8 |
Hb_004435_020 |
0.1387507691 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 9 |
Hb_007218_040 |
0.1422602171 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 10 |
Hb_001140_300 |
0.1441933741 |
- |
- |
PREDICTED: tubulin beta-1 chain [Cicer arietinum] |
| 11 |
Hb_007632_070 |
0.1452493461 |
- |
- |
kinase, putative [Ricinus communis] |
| 12 |
Hb_000003_750 |
0.1458990372 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 13 |
Hb_010889_010 |
0.1464390814 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 14 |
Hb_000735_020 |
0.1466122533 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 15 |
Hb_003097_130 |
0.1476616063 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_010381_100 |
0.1495331607 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 17 |
Hb_007811_020 |
0.1523534758 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 18 |
Hb_001287_090 |
0.1535780415 |
- |
- |
Endoglucanase 4 [Aegilops tauschii] |
| 19 |
Hb_005675_050 |
0.1536272816 |
- |
- |
alpha-glucosidase, putative [Ricinus communis] |
| 20 |
Hb_002045_210 |
0.1548606397 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |