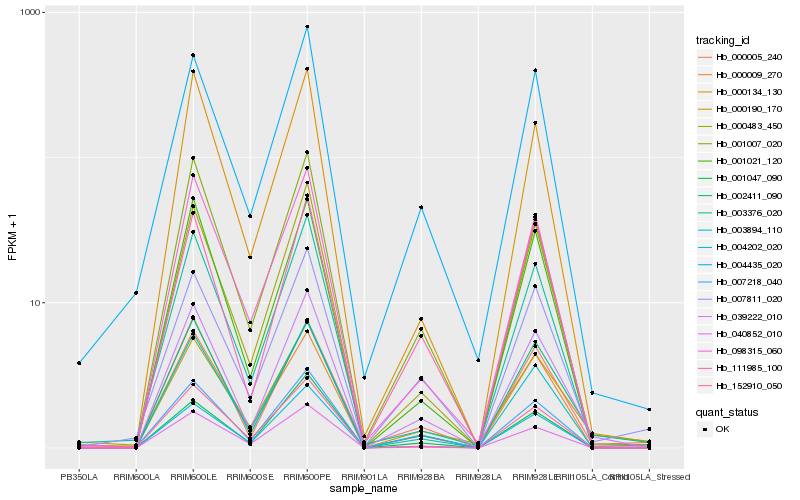
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039222_010 |
0.0 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003894_110 |
0.0855082702 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |
| 3 |
Hb_000005_240 |
0.0862630724 |
- |
- |
Shugoshin-1, putative [Ricinus communis] |
| 4 |
Hb_007218_040 |
0.0862717235 |
- |
- |
Leucine-rich repeat transmembrane protein kinase protein, putative [Theobroma cacao] |
| 5 |
Hb_000483_450 |
0.0893289859 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 6 |
Hb_002411_090 |
0.0895631954 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL69 [Jatropha curcas] |
| 7 |
Hb_001021_120 |
0.0934444961 |
transcription factor |
TF Family: NF-YB |
PREDICTED: nuclear transcription factor Y subunit B-4 [Jatropha curcas] |
| 8 |
Hb_000009_270 |
0.0938091043 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 9 |
Hb_000190_170 |
0.095533121 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 10 |
Hb_000134_130 |
0.0983573793 |
- |
- |
PREDICTED: uncharacterized protein LOC105628670 [Jatropha curcas] |
| 11 |
Hb_098315_060 |
0.098577421 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 12 |
Hb_111985_100 |
0.1009510391 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 13 |
Hb_004202_020 |
0.1023654274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_152910_050 |
0.1032627789 |
transcription factor |
TF Family: OFP |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004435_020 |
0.1032950964 |
- |
- |
PREDICTED: tubulin alpha-2 chain [Jatropha curcas] |
| 16 |
Hb_007811_020 |
0.1045585827 |
- |
- |
PREDICTED: expansin-A6 [Jatropha curcas] |
| 17 |
Hb_040852_010 |
0.1052622232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001007_020 |
0.1061042213 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003376_020 |
0.1069559651 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_001047_090 |
0.1070121237 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |