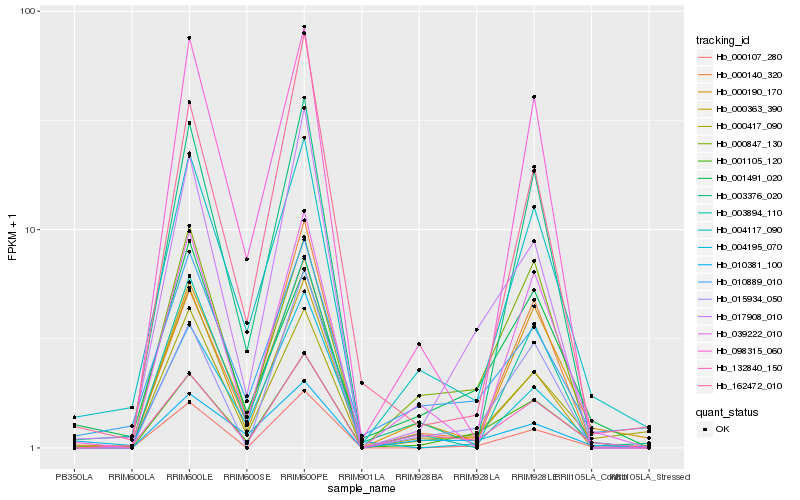
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001105_120 |
0.0 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 2 |
Hb_132840_150 |
0.1274353733 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 3 |
Hb_017908_010 |
0.1349717013 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000417_090 |
0.1497171234 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000190_170 |
0.154249754 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 6 |
Hb_004117_090 |
0.1549759941 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 70 [Jatropha curcas] |
| 7 |
Hb_010381_100 |
0.1562986903 |
- |
- |
Phospholipase D epsilon [Morus notabilis] |
| 8 |
Hb_015934_050 |
0.1604389409 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 9 |
Hb_000107_280 |
0.1620528378 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000363_390 |
0.1644688108 |
- |
- |
Benzoate carboxyl methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_010889_010 |
0.1649982624 |
- |
- |
PREDICTED: myosin-12 [Jatropha curcas] |
| 12 |
Hb_000847_130 |
0.1679248006 |
- |
- |
PREDICTED: fimbrin-2 [Jatropha curcas] |
| 13 |
Hb_039222_010 |
0.171181896 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003376_020 |
0.175657931 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000140_320 |
0.1771275458 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 16 |
Hb_001491_020 |
0.1778331383 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_098315_060 |
0.1801214239 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 7-like [Jatropha curcas] |
| 18 |
Hb_162472_010 |
0.1802543155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004195_070 |
0.1802695942 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_003894_110 |
0.180527745 |
- |
- |
PREDICTED: 65-kDa microtubule-associated protein 4 [Jatropha curcas] |