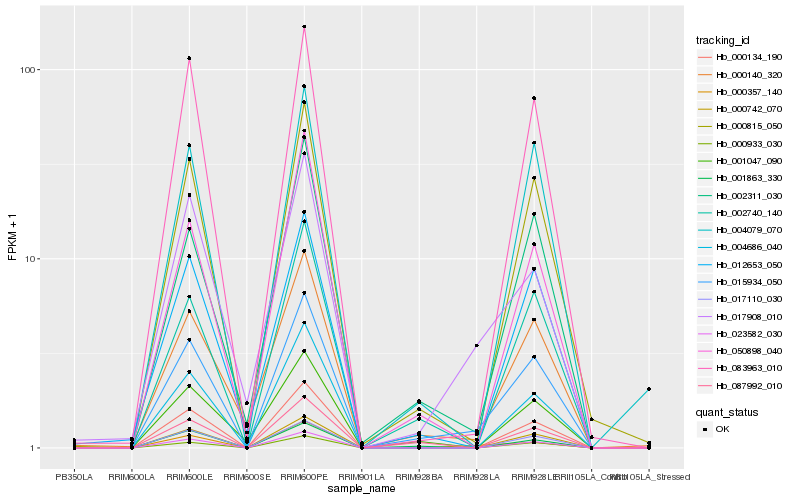
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015934_050 |
0.0 |
- |
- |
PREDICTED: transcription factor PAR2-like [Jatropha curcas] |
| 2 |
Hb_000815_050 |
0.1143073691 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 3 |
Hb_000140_320 |
0.1199764646 |
- |
- |
delta-8 sphingolipid desaturase [Vernicia fordii] |
| 4 |
Hb_002740_140 |
0.1203231418 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 5 |
Hb_002311_030 |
0.1227524176 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_023582_030 |
0.1296568833 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 7 |
Hb_000742_070 |
0.1311823816 |
- |
- |
PREDICTED: monoglyceride lipase isoform X2 [Bubalus bubalis] |
| 8 |
Hb_000134_190 |
0.1317513501 |
- |
- |
hypothetical protein JCGZ_16725 [Jatropha curcas] |
| 9 |
Hb_087992_010 |
0.1340023703 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 10 |
Hb_000933_030 |
0.1340316549 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004079_070 |
0.1351014573 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 12 |
Hb_083963_010 |
0.1357679152 |
- |
- |
PREDICTED: probable carboxylesterase 8 [Jatropha curcas] |
| 13 |
Hb_017908_010 |
0.1360774809 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_017110_030 |
0.1363548265 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.4 [Jatropha curcas] |
| 15 |
Hb_012653_050 |
0.1376253267 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 16 |
Hb_000357_140 |
0.1385587851 |
- |
- |
hypothetical protein JCGZ_15089 [Jatropha curcas] |
| 17 |
Hb_004686_040 |
0.1392166665 |
- |
- |
hypothetical protein POPTR_0016s05690g [Populus trichocarpa] |
| 18 |
Hb_050898_040 |
0.1405611268 |
- |
- |
- |
| 19 |
Hb_001863_330 |
0.1408987879 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 20 |
Hb_001047_090 |
0.1421296354 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP16-like [Jatropha curcas] |