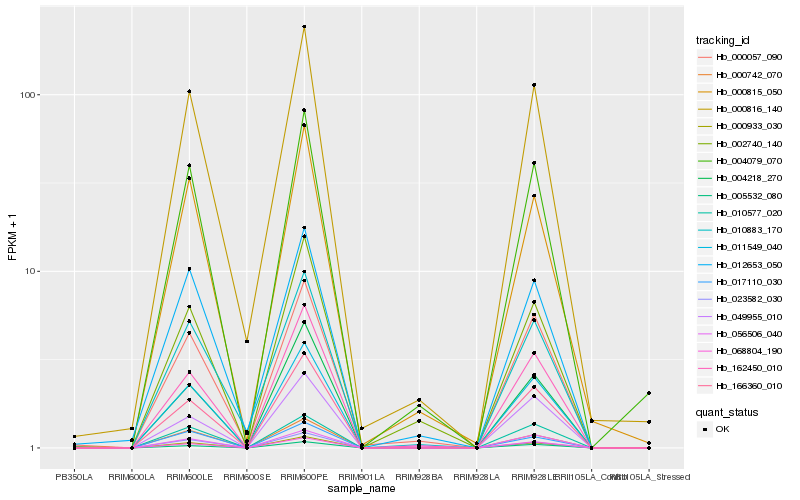
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004079_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 2 |
Hb_011549_040 |
0.0771838092 |
- |
- |
- |
| 3 |
Hb_000933_030 |
0.0795117025 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000815_050 |
0.081462517 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 5 |
Hb_000816_140 |
0.0841513005 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 6 |
Hb_068804_190 |
0.0899031247 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_166360_010 |
0.0906463296 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 8 |
Hb_000742_070 |
0.0910500063 |
- |
- |
PREDICTED: monoglyceride lipase isoform X2 [Bubalus bubalis] |
| 9 |
Hb_000057_090 |
0.0917256657 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 10 |
Hb_162450_010 |
0.0931294615 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 11 |
Hb_010577_020 |
0.0945450724 |
- |
- |
- |
| 12 |
Hb_002740_140 |
0.0947671346 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 13 |
Hb_012653_050 |
0.0975246327 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 14 |
Hb_005532_080 |
0.0987306691 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 15 |
Hb_023582_030 |
0.0992473438 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 16 |
Hb_056506_040 |
0.1008688442 |
- |
- |
- |
| 17 |
Hb_010883_170 |
0.1026635204 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 18 |
Hb_017110_030 |
0.1043949986 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.4 [Jatropha curcas] |
| 19 |
Hb_004218_270 |
0.1077363807 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |
| 20 |
Hb_049955_010 |
0.1081885975 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |