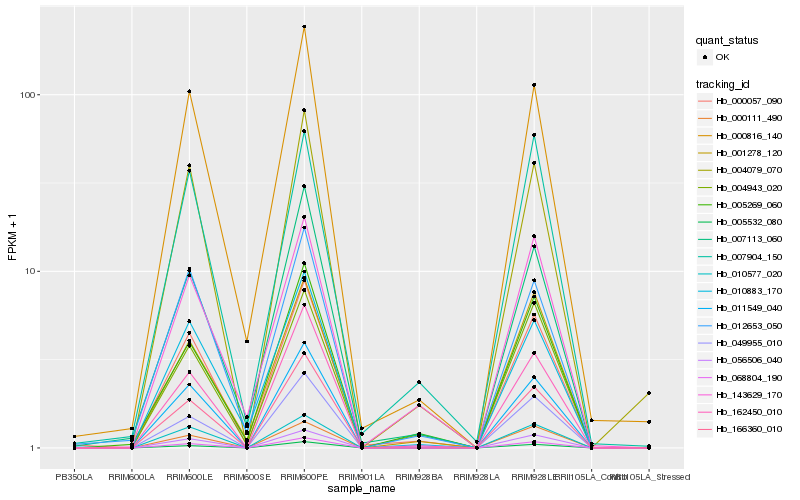
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000057_090 |
0.0 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 2 |
Hb_000816_140 |
0.0756217143 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 3 |
Hb_068804_190 |
0.0805321635 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_011549_040 |
0.0839680418 |
- |
- |
- |
| 5 |
Hb_049955_010 |
0.0842673049 |
- |
- |
hypothetical protein EUGRSUZ_B00742 [Eucalyptus grandis] |
| 6 |
Hb_056506_040 |
0.085601261 |
- |
- |
- |
| 7 |
Hb_010883_170 |
0.0857165495 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 8 |
Hb_005532_080 |
0.0868184911 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 9 |
Hb_007113_060 |
0.0874549469 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 10 |
Hb_166360_010 |
0.0890839502 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 11 |
Hb_004943_020 |
0.0900193409 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_012653_050 |
0.0910936783 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 13 |
Hb_162450_010 |
0.0912142836 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 14 |
Hb_004079_070 |
0.0917256657 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 15 |
Hb_010577_020 |
0.0927195273 |
- |
- |
- |
| 16 |
Hb_001278_120 |
0.0938610196 |
- |
- |
PREDICTED: reticulon-like protein B14 [Jatropha curcas] |
| 17 |
Hb_005269_060 |
0.0950781419 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Cicer arietinum] |
| 18 |
Hb_143629_170 |
0.096491636 |
- |
- |
PREDICTED: pectin acetylesterase 12 [Jatropha curcas] |
| 19 |
Hb_007904_150 |
0.0979445468 |
- |
- |
PREDICTED: probable pectate lyase 5 [Populus euphratica] |
| 20 |
Hb_000111_490 |
0.0989187942 |
- |
- |
conserved hypothetical protein [Ricinus communis] |