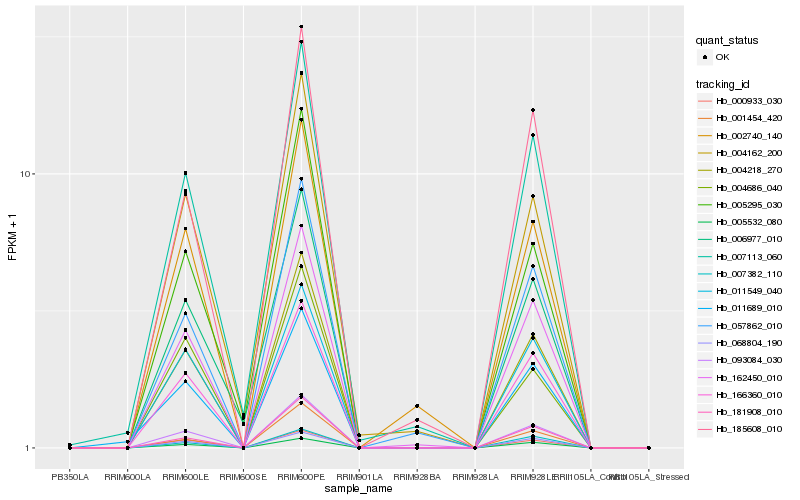
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_270 |
0.0 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |
| 2 |
Hb_093084_030 |
0.0185992695 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 3 |
Hb_162450_010 |
0.0464943183 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_166360_010 |
0.0477148745 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 5 |
Hb_000933_030 |
0.0600320751 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011549_040 |
0.0672919543 |
- |
- |
- |
| 7 |
Hb_005532_080 |
0.0690692787 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 8 |
Hb_068804_190 |
0.0791965704 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_057862_010 |
0.0809917084 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_185608_010 |
0.0830749713 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 11 |
Hb_005295_030 |
0.0858931454 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 12 |
Hb_007113_060 |
0.0878323355 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 13 |
Hb_011689_010 |
0.0879013465 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 14 |
Hb_181908_010 |
0.0884571294 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 15 |
Hb_006977_010 |
0.0919212798 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001454_420 |
0.091962418 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 17 |
Hb_004162_200 |
0.0924095642 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 18 |
Hb_007382_110 |
0.0934798196 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 19 |
Hb_002740_140 |
0.0940143037 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 20 |
Hb_004686_040 |
0.0952605415 |
- |
- |
hypothetical protein POPTR_0016s05690g [Populus trichocarpa] |