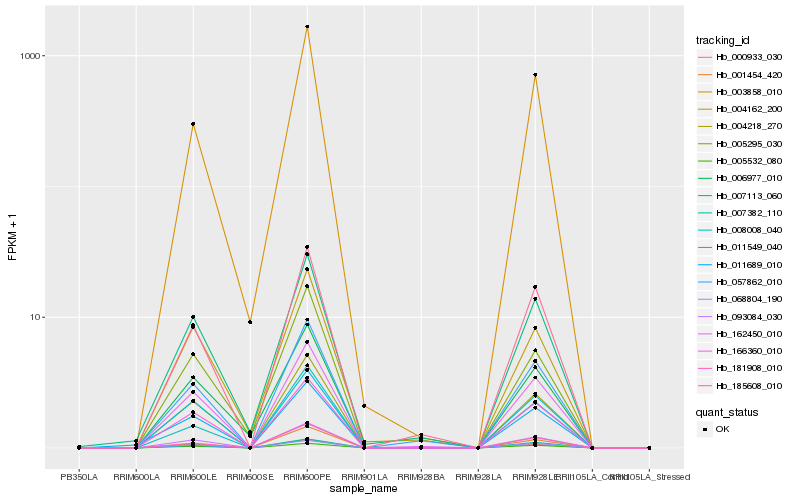
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_093084_030 |
0.0 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 2 |
Hb_004218_270 |
0.0185992695 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |
| 3 |
Hb_162450_010 |
0.048347618 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_166360_010 |
0.0566052774 |
- |
- |
hypothetical protein POPTR_0010s12130g [Populus trichocarpa] |
| 5 |
Hb_181908_010 |
0.0699743469 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 6 |
Hb_185608_010 |
0.0728665922 |
- |
- |
hypothetical protein POPTR_0004s22210g [Populus trichocarpa] |
| 7 |
Hb_005532_080 |
0.0737892969 |
- |
- |
PREDICTED: heparanase-like protein 2 [Jatropha curcas] |
| 8 |
Hb_057862_010 |
0.0738924104 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_001454_420 |
0.0739093179 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 10 |
Hb_000933_030 |
0.0782205753 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003858_010 |
0.0801529143 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 12 |
Hb_011549_040 |
0.0816125315 |
- |
- |
- |
| 13 |
Hb_005295_030 |
0.084236996 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 14 |
Hb_007382_110 |
0.0872685577 |
- |
- |
PREDICTED: bidirectional sugar transporter N3 [Jatropha curcas] |
| 15 |
Hb_008008_040 |
0.0873126424 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 16 |
Hb_068804_190 |
0.0878330561 |
- |
- |
kinase, putative [Ricinus communis] |
| 17 |
Hb_007113_060 |
0.0890746182 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 18 |
Hb_011689_010 |
0.0915268919 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 19 |
Hb_006977_010 |
0.0944641766 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_004162_200 |
0.0979630517 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |