| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
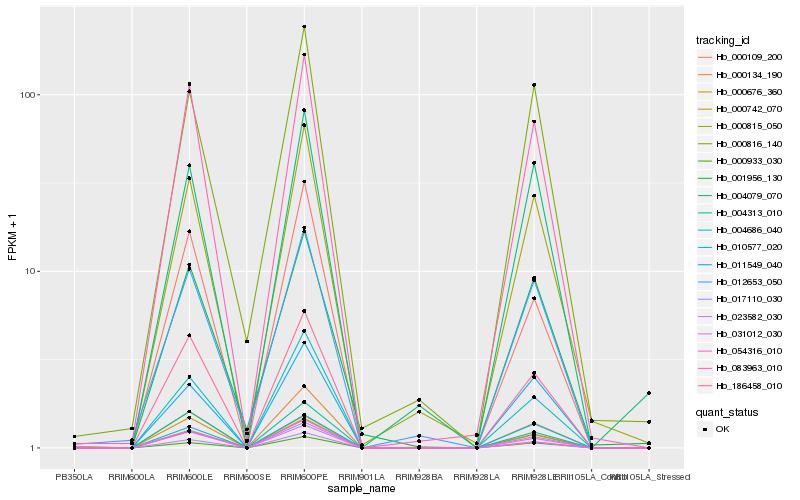
Hb_017110_030 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.4 [Jatropha curcas] |
| 2 |
Hb_000742_070 |
0.0245367115 |
- |
- |
PREDICTED: monoglyceride lipase isoform X2 [Bubalus bubalis] |
| 3 |
Hb_083963_010 |
0.0341226781 |
- |
- |
PREDICTED: probable carboxylesterase 8 [Jatropha curcas] |
| 4 |
Hb_186458_010 |
0.0349873605 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 5 |
Hb_023582_030 |
0.0350326077 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 6 |
Hb_000134_190 |
0.0513563123 |
- |
- |
hypothetical protein JCGZ_16725 [Jatropha curcas] |
| 7 |
Hb_000933_030 |
0.0701383272 |
transcription factor |
TF Family: ERF |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_004313_010 |
0.0728731816 |
- |
- |
PREDICTED: patatin-like protein 1 [Prunus mume] |
| 9 |
Hb_004686_040 |
0.0840798888 |
- |
- |
hypothetical protein POPTR_0016s05690g [Populus trichocarpa] |
| 10 |
Hb_000815_050 |
0.0844233072 |
- |
- |
PREDICTED: expansin-A1 [Populus euphratica] |
| 11 |
Hb_031012_030 |
0.0852995251 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT5-like [Populus euphratica] |
| 12 |
Hb_011549_040 |
0.0930572993 |
- |
- |
- |
| 13 |
Hb_012653_050 |
0.0983573719 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 14 |
Hb_000676_360 |
0.0995270188 |
- |
- |
hypothetical protein POPTR_0007s13690g [Populus trichocarpa] |
| 15 |
Hb_001956_130 |
0.1035078104 |
- |
- |
catalytic, putative [Ricinus communis] |
| 16 |
Hb_000109_200 |
0.1041083241 |
- |
- |
hypothetical protein CISIN_1g037825mg [Citrus sinensis] |
| 17 |
Hb_004079_070 |
0.1043949986 |
- |
- |
hypothetical protein JCGZ_07230 [Jatropha curcas] |
| 18 |
Hb_010577_020 |
0.1045400278 |
- |
- |
- |
| 19 |
Hb_054316_010 |
0.1110772078 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 20 |
Hb_000816_140 |
0.1139546773 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |