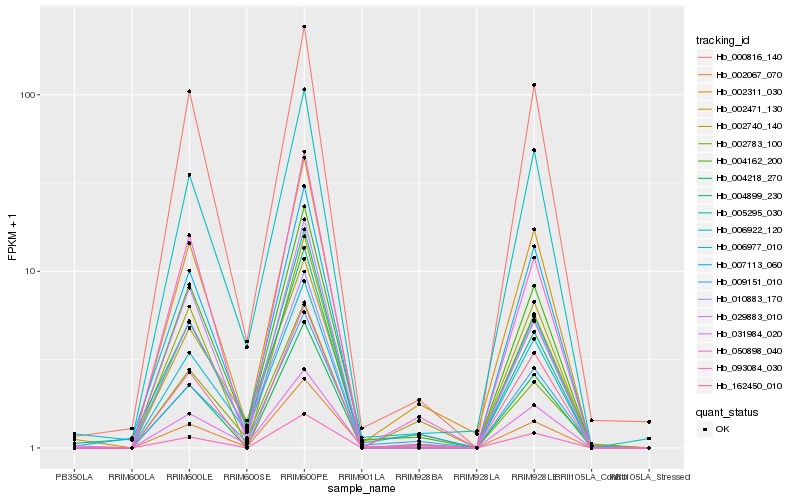
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004162_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At1g08160 [Jatropha curcas] |
| 2 |
Hb_002067_070 |
0.0622881351 |
- |
- |
hypothetical protein JCGZ_19254 [Jatropha curcas] |
| 3 |
Hb_002311_030 |
0.0638679485 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_007113_060 |
0.0702891413 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 5 |
Hb_005295_030 |
0.0733328372 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 6 |
Hb_009151_010 |
0.0739432723 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 7 |
Hb_050898_040 |
0.0769955356 |
- |
- |
- |
| 8 |
Hb_006977_010 |
0.079357608 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_000816_140 |
0.0808665198 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 10 |
Hb_006922_120 |
0.0830863228 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 11 |
Hb_002783_100 |
0.0878384226 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 12 |
Hb_010883_170 |
0.0910252286 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 13 |
Hb_031984_020 |
0.092223876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004218_270 |
0.0924095642 |
- |
- |
hypothetical protein CICLE_v100134152mg, partial [Citrus clementina] |
| 15 |
Hb_029883_010 |
0.0927582764 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 16 |
Hb_004899_230 |
0.0940065052 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 17 |
Hb_002740_140 |
0.0948107266 |
- |
- |
hypothetical protein L484_022679 [Morus notabilis] |
| 18 |
Hb_162450_010 |
0.095386756 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 19 |
Hb_093084_030 |
0.0979630517 |
- |
- |
oleosin2 [Plukenetia volubilis] |
| 20 |
Hb_002471_130 |
0.0989501832 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 3 [Jatropha curcas] |