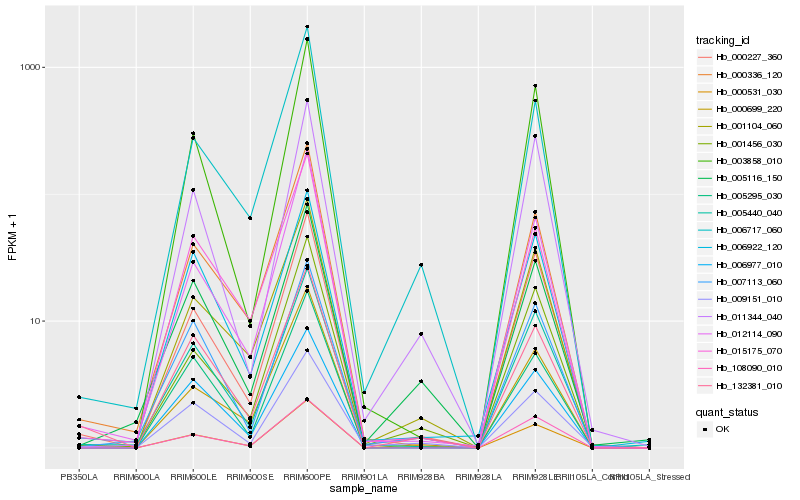
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000531_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001104_060 |
0.0539067444 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9 [Jatropha curcas] |
| 3 |
Hb_005440_040 |
0.0744017847 |
- |
- |
PREDICTED: microtubule-associated protein 70-5 [Jatropha curcas] |
| 4 |
Hb_000227_360 |
0.0745997291 |
- |
- |
PREDICTED: mavicyanin [Jatropha curcas] |
| 5 |
Hb_009151_010 |
0.08182042 |
- |
- |
Peroxidase 3 [Morus notabilis] |
| 6 |
Hb_001456_030 |
0.0829314376 |
- |
- |
PREDICTED: bifunctional pinoresinol-lariciresinol reductase 2 [Vitis vinifera] |
| 7 |
Hb_005116_150 |
0.0840833433 |
- |
- |
PREDICTED: cyclin-P3-1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003858_010 |
0.0841538743 |
- |
- |
xyloglucan endotransglucosylase/hydrolase [Gossypium hirsutum] |
| 9 |
Hb_000699_220 |
0.0842627673 |
- |
- |
PREDICTED: phospholipid-metabolizing enzyme A-C1-like [Jatropha curcas] |
| 10 |
Hb_006717_060 |
0.0845432247 |
- |
- |
PREDICTED: peroxidase 42 [Jatropha curcas] |
| 11 |
Hb_108090_010 |
0.0854554343 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_006977_010 |
0.087364037 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_011344_040 |
0.0880205697 |
- |
- |
PREDICTED: 21 kDa protein [Jatropha curcas] |
| 14 |
Hb_000336_120 |
0.0887385907 |
- |
- |
PREDICTED: mavicyanin [Populus euphratica] |
| 15 |
Hb_007113_060 |
0.0902324036 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 16 |
Hb_006922_120 |
0.0922540038 |
- |
- |
hypothetical protein JCGZ_01146 [Jatropha curcas] |
| 17 |
Hb_012114_090 |
0.0927160976 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 18 |
Hb_005295_030 |
0.0946688197 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 19 |
Hb_015175_070 |
0.0951683951 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 20 |
Hb_132381_010 |
0.0954705041 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |